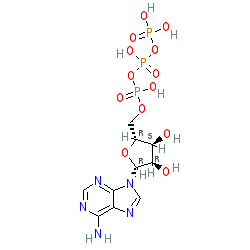
|
Synonyms: 5'-ATP | adenosine 5' triphosphate | adenosine triphosphate
Compound class:
Metabolite or derivative
Ligand Activity Visualisation ChartsThese are box plot that provide a unique visualisation, summarising all the activity data for a ligand taken from ChEMBL and GtoPdb across multiple targets and species. Click on a plot to see the median, interquartile range, low and high data points. A value of zero indicates that no data are available. A separate chart is created for each target, and where possible the algorithm tries to merge ChEMBL and GtoPdb targets by matching them on name and UniProt accession, for each available species. However, please note that inconsistency in naming of targets may lead to data for the same target being reported across multiple charts. ✖
View more information in the IUPHAR Pharmacology Education Project: atp |
|
|||||||||||||||||||||||||||||||||||
| Natural/Endogenous Targets | |||||||||||||||
|
|||||||||||||||
| Enzymes Catalysing Reactions with this Compound as a Substrate or Product | ||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||
| Cofactor in Enzyme Reactions | ||||
|
||||
| Transporters Moving this Compound Across a Lipid Membrane | ||||||||||||
|
||||||||||||
| Selectivity at GPCRs | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Selectivity at ion channels | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | Click column headers to sort | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Additional information and targets (data relate to human unless otherwise stated) | ||
| Description | Data | Reference |
| Potency order of endogenous ligands at P2Y12 receptor | ADP>ATP | |
| Other channel blockers at TRPM4 | Intracellular nucleotides including ATP, ADP, adenosine 5'-monophosphate and AMP-PNP with an IC50 range of 1.3-1.9 μM | |
| Potency order of endogenous ligands at P2Y13 receptor | ADP>>ATP | |
| Potency order of endogenous ligands at P2Y1 receptor | ADP>ATP | |
| Potency order of endogenous ligands at P2Y4 receptor | UTP>ATP (at rat recombinant receptors, UTP = ATP) | |
| Potency order of endogenous ligands at P2Y11 receptor | ATP>ADP | |
| Potency order of endogenous ligands at P2Y2 receptor | UTP > ATP | |
| Targets where the ligand is described in the comment field | |
| Target | Comment |
| TRPM5 | TRPM5 is not blocked by ATP. APV206512A and APV206513A are TRMP5 blockers, with IC50s of 15 μM [35]. Steviol glycosides (sweet-tasting organic molecules) act as positive modulators of TRMP5 activity [26]. |
| Ligand mentioned in the following text fields |
| Adenosine turnover overview |
| Adenylyl cyclases (ACs) overview |
| F-type ATPase overview |
| F-type and V-type ATPases overview |
| Kinases (EC 2.7.x.x) overview |
| Maxi chloride channel overview |
| Mitochondrial nucleotide transporter subfamily overview |
| Mitochondrial phosphate transporters overview |
| P2Y receptors overview |
| V-type ATPase overview |
| Vesicular nucleotide transporter overview |
| CFTR comments |
| Maxi chloride channel comments |
| NOD-like receptor family comments |
| Phosphatidylcholine-specific phospholipase D comments |
| SLC33 acetylCoA transporter comments |