GtoPdb is requesting financial support from commercial users. Please see our sustainability page for more information.
oxidative stress responsive kinase 1
Target not currently curated in GtoImmuPdb
Target id: 2132
Nomenclature: oxidative stress responsive kinase 1
Abbreviated Name: OSR1
Family: FRAY subfamily
Contents:
Gene and Protein Information  |
||||||
| Species | TM | AA | Chromosomal Location | Gene Symbol | Gene Name | Reference |
| Human | - | 527 | 3p22.2 | OXSR1 | oxidative stress responsive kinase 1 | |
| Mouse | - | 527 | 9 F3 | Oxsr1 | oxidative-stress responsive 1 | |
| Rat | - | 527 | 8 q32 | Oxsr1 | oxidative stress responsive kinase 1 | |
Previous and Unofficial Names  |
| oxidative stress responsive 1 |
Database Links  |
|
| Alphafold | O95747 (Hs), Q6P9R2 (Mm) |
| BRENDA | 2.7.11.1 |
| ChEMBL Target | CHEMBL1163104 (Hs) |
| Ensembl Gene | ENSG00000172939 (Hs), ENSMUSG00000036737 (Mm), ENSRNOG00000013136 (Rn) |
| Entrez Gene | 9943 (Hs), 108737 (Mm), 316064 (Rn) |
| Human Protein Atlas | ENSG00000172939 (Hs) |
| KEGG Enzyme | 2.7.11.1 |
| KEGG Gene | hsa:9943 (Hs), mmu:108737 (Mm), rno:316064 (Rn) |
| OMIM | 604046 (Hs) |
| Pharos | O95747 (Hs) |
| RefSeq Nucleotide | NM_005109 (Hs), NM_133985 (Mm), NM_001108194 (Rn) |
| RefSeq Protein | NP_005100 (Hs), NP_598746 (Mm), NP_001101664 (Rn) |
| UniProtKB | O95747 (Hs), Q6P9R2 (Mm) |
| Wikipedia | OXSR1 (Hs) |
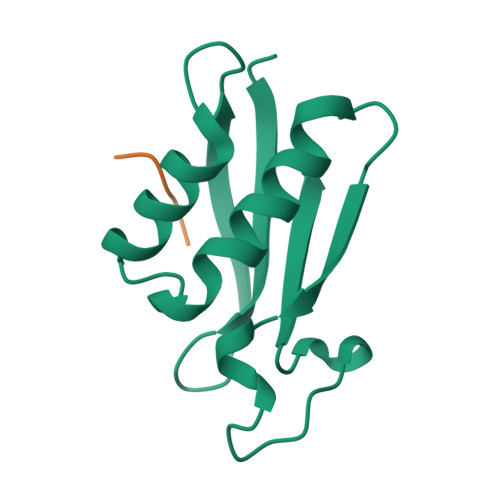
Selected 3D Structures  |
|||||||||||
|
|
||||||||||
Enzyme Reaction  |
||||
|
||||
Download all structure-activity data for this target as a CSV file 
| Inhibitors | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | View all chemical structures | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| Inhibitor Comments | |||||||||||||||||||||||||||||||||||||||||||||||||||
| OXSR1 activity is inhibited by approximately 86% (average of n=2) in the presence of 1μM compound 74 [PMID 24793884] [3] (note that OXSR1 is referred to as OSR1 in the article's Supplementary data table).. | |||||||||||||||||||||||||||||||||||||||||||||||||||
DiscoveRx KINOMEscan® screen  |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A screen of 72 inhibitors against 456 human kinases. Quantitative data were derived using DiscoveRx KINOMEscan® platform. http://www.discoverx.com/services/drug-discovery-development-services/kinase-profiling/kinomescan Reference: 2,5 |
 
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | Click column headers to sort | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Target used in screen: OSR1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displaying the top 10 most potent ligands View all ligands in screen » | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
EMD Millipore KinaseProfilerTM screen/Reaction Biology Kinase HotspotSM screen  |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A screen profiling 158 kinase inhibitors (Calbiochem Protein Kinase Inhibitor Library I and II, catalogue numbers 539744 and 539745) for their inhibitory activity at 1µM and 10µM against 234 human recombinant kinases using the EMD Millipore KinaseProfilerTM service. A screen profiling the inhibitory activity of 178 commercially available kinase inhibitors at 0.5µM against a panel of 300 recombinant protein kinases using the Reaction Biology Corporation Kinase HotspotSM platform. http://www.millipore.com/techpublications/tech1/pf3036 http://www.reactionbiology.com/webapps/main/pages/kinase.aspx Reference: ...1 |
 
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Target used in screen: nd/OSR1(OXSR1) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displaying the top 10 most potent ligands View all ligands in screen » | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
References
1. Anastassiadis T, Deacon SW, Devarajan K, Ma H, Peterson JR. (2011) Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nat Biotechnol, 29 (11): 1039-45. [PMID:22037377]
2. Davis MI, Hunt JP, Herrgard S, Ciceri P, Wodicka LM, Pallares G, Hocker M, Treiber DK, Zarrinkar PP. (2011) Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol, 29 (11): 1046-51. [PMID:22037378]
3. Reichelt A, Bailis JM, Bartberger MD, Yao G, Shu H, Kaller MR, Allen JG, Weidner MF, Keegan KS, Dao JH. (2014) Synthesis and structure-activity relationship of trisubstituted thiazoles as Cdc7 kinase inhibitors. Eur J Med Chem, 80: 364-82. [PMID:24793884]
4. Villa F, Goebel J, Rafiqi FH, Deak M, Thastrup J, Alessi DR, van Aalten DM. (2007) Structural insights into the recognition of substrates and activators by the OSR1 kinase. EMBO Rep, 8 (9): 839-45. [PMID:17721439]
5. Wodicka LM, Ciceri P, Davis MI, Hunt JP, Floyd M, Salerno S, Hua XH, Ford JM, Armstrong RC, Zarrinkar PP et al.. (2010) Activation state-dependent binding of small molecule kinase inhibitors: structural insights from biochemistry. Chem Biol, 17 (11): 1241-9. [PMID:21095574]
How to cite this page
FRAY subfamily: oxidative stress responsive kinase 1. Last modified on 27/03/2019. Accessed on 31/01/2026. IUPHAR/BPS Guide to PHARMACOLOGY, https://www.guidetoimmunopharmacology.org/GRAC/ObjectDisplayForward?objectId=2132.