GtoPdb is requesting financial support from commercial users. Please see our sustainability page for more information.
mitogen-activated protein kinase 12
 Target has curated data in GtoImmuPdb
Target has curated data in GtoImmuPdb
Target id: 1501
Nomenclature: mitogen-activated protein kinase 12
Abbreviated Name: p38γ
Family: p38 subfamily
Contents:
Gene and Protein Information  |
||||||
| Species | TM | AA | Chromosomal Location | Gene Symbol | Gene Name | Reference |
| Human | - | 367 | 22q13.33 | MAPK12 | mitogen-activated protein kinase 12 | |
| Mouse | - | 367 | 15 E3 | Mapk12 | mitogen-activated protein kinase 12 | |
| Rat | - | 367 | 7q34 | Mapk12 | mitogen-activated protein kinase 12 | |
Database Links  |
|
| Alphafold | P53778 (Hs), O08911 (Mm), Q63538 (Rn) |
| BRENDA | 2.7.11.24 |
| ChEMBL Target | CHEMBL4674 (Hs) |
| Ensembl Gene | ENSG00000188130 (Hs), ENSMUSG00000022610 (Mm), ENSRNOG00000031233 (Rn) |
| Entrez Gene | 6300 (Hs), 29857 (Mm), 60352 (Rn) |
| Human Protein Atlas | ENSG00000188130 (Hs) |
| KEGG Enzyme | 2.7.11.24 |
| KEGG Gene | hsa:6300 (Hs), mmu:29857 (Mm), rno:60352 (Rn) |
| OMIM | 602399 (Hs) |
| Pharos | P53778 (Hs) |
| RefSeq Nucleotide | XM_003846644 (Hs), NM_013871 (Mm), NM_021746 (Rn) |
| RefSeq Protein | NP_002960 (Hs), NP_038899 (Mm), NP_068514 (Rn) |
| UniProtKB | P53778 (Hs), O08911 (Mm), Q63538 (Rn) |
| Wikipedia | MAPK12 (Hs) |
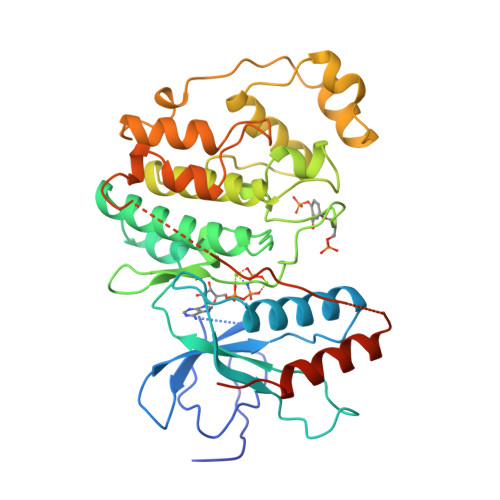
Selected 3D Structures  |
|||||||||||
|
|
||||||||||
Enzyme Reaction  |
||||
|
||||
Download all structure-activity data for this target as a CSV file 
| Inhibitors | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | View all chemical structures | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
DiscoveRx KINOMEscan® screen  |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A screen of 72 inhibitors against 456 human kinases. Quantitative data were derived using DiscoveRx KINOMEscan® platform. http://www.discoverx.com/services/drug-discovery-development-services/kinase-profiling/kinomescan Reference: 5,14 |
 
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | Click column headers to sort | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Target used in screen: p38-gamma | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displaying the top 10 most potent ligands View all ligands in screen » | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
EMD Millipore KinaseProfilerTM screen/Reaction Biology Kinase HotspotSM screen  |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A screen profiling 158 kinase inhibitors (Calbiochem Protein Kinase Inhibitor Library I and II, catalogue numbers 539744 and 539745) for their inhibitory activity at 1µM and 10µM against 234 human recombinant kinases using the EMD Millipore KinaseProfilerTM service. A screen profiling the inhibitory activity of 178 commercially available kinase inhibitors at 0.5µM against a panel of 300 recombinant protein kinases using the Reaction Biology Corporation Kinase HotspotSM platform. http://www.millipore.com/techpublications/tech1/pf3036 http://www.reactionbiology.com/webapps/main/pages/kinase.aspx Reference: 1,6 |
 
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Target used in screen: SAPK3/P38g | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displaying the top 10 most potent ligands View all ligands in screen » | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Immunopharmacology Comments |
| p38 MAP kinases are ubiquitous, highly conserved enzymes which regulate the production of proinflammatory mediators (such as TNFα and IL-1) in response to inflammatory cytokines or environmental stress [7-9,11-13]. They are essential for normal immune and inflammatory responses, but are also involved in many other cellular processes such as regulating the cell cycle and cytoskeletal remodelling. Pharmacological inhibition of p38 MAP kinases reduces inflammatory cytokine synthesis, making these enzymes validated and extensively pursued drug targets for autoimmune and inflammatory diseases, including arthritis and other joint diseases, septic shock, myocardial injury and neuroinflammation. A number of pan-p38 MAP kinase inhibitors and isoform selective inhibitors have been evaluated in clinical trials, although none have yet reached the clinic. |
| General Comments |
| SARS-CoV-2: p38 MAP kinase activity is reported to be upregulated by SARS-CoV-2 infection in vitro [4]. siRNA-mediated knockdown of MAPK12 (p38γ) in A549-ACE2 cells significantly reduces SARS-CoV-2 replication, without reducing cell viability. |
References
1. Anastassiadis T, Deacon SW, Devarajan K, Ma H, Peterson JR. (2011) Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nat Biotechnol, 29 (11): 1039-45. [PMID:22037377]
2. Armani E, Capaldi C, Bagnacani V, Saccani F, Aquino G, Puccini P, Facchinetti F, Martucci C, Moretto N, Villetti G et al.. (2022) Design, Synthesis, and Biological Characterization of Inhaled p38α/β MAPK Inhibitors for the Treatment of Lung Inflammatory Diseases. J Med Chem, 65 (10): 7170-7192. [PMID:35546685]
3. Bellon S, Fitzgibbon MJ, Fox T, Hsiao HM, Wilson KP. (1999) The structure of phosphorylated p38gamma is monomeric and reveals a conserved activation-loop conformation. Structure, 7 (9): 1057-65. [PMID:10508788]
4. Bouhaddou M, Memon D, Meyer B, White KM, Rezelj VV, Marrero MC, Polacco BJ, Melnyk JE, Ulferts S, Kaake RM. (2020) The Global Phosphorylation Landscape of SARS-CoV-2 Infection. Cell, Article Online Now. DOI: 10.1016/j.cell.2020.06.034
5. Davis MI, Hunt JP, Herrgard S, Ciceri P, Wodicka LM, Pallares G, Hocker M, Treiber DK, Zarrinkar PP. (2011) Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol, 29 (11): 1046-51. [PMID:22037378]
6. Gao Y, Davies SP, Augustin M, Woodward A, Patel UA, Kovelman R, Harvey KJ. (2013) A broad activity screen in support of a chemogenomic map for kinase signalling research and drug discovery. Biochem J, 451 (2): 313-28. [PMID:23398362]
7. Han J, Jiang Y, Li Z, Kravchenko VV, Ulevitch RJ. (1997) Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature, 386 (6622): 296-9. [PMID:9069290]
8. Han J, Lee JD, Bibbs L, Ulevitch RJ. (1994) A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cells. Science, 265 (5173): 808-11. [PMID:7914033]
9. Lee JC, Kumar S, Griswold DE, Underwood DC, Votta BJ, Adams JL. (2000) Inhibition of p38 MAP kinase as a therapeutic strategy. Immunopharmacology, 47 (2-3): 185-201. [PMID:10878289]
10. Moffett K, Konteatis Z, Nguyen D, Shetty R, Ludington J, Fujimoto T, Lee KJ, Chai X, Namboodiri H, Karpusas M et al.. (2011) Discovery of a novel class of non-ATP site DFG-out state p38 inhibitors utilizing computationally assisted virtual fragment-based drug design (vFBDD). Bioorg Med Chem Lett, 21 (23): 7155-65. [PMID:22014550]
11. Pearson G, Robinson F, Beers Gibson T, Xu BE, Karandikar M, Berman K, Cobb MH. (2001) Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr Rev, 22 (2): 153-83. [PMID:11294822]
12. Raingeaud J, Gupta S, Rogers JS, Dickens M, Han J, Ulevitch RJ, Davis RJ. (1995) Pro-inflammatory cytokines and environmental stress cause p38 mitogen-activated protein kinase activation by dual phosphorylation on tyrosine and threonine. J Biol Chem, 270 (13): 7420-6. [PMID:7535770]
13. Wang XZ, Ron D. (1996) Stress-induced phosphorylation and activation of the transcription factor CHOP (GADD153) by p38 MAP Kinase. Science, 272 (5266): 1347-9. [PMID:8650547]
14. Wodicka LM, Ciceri P, Davis MI, Hunt JP, Floyd M, Salerno S, Hua XH, Ford JM, Armstrong RC, Zarrinkar PP et al.. (2010) Activation state-dependent binding of small molecule kinase inhibitors: structural insights from biochemistry. Chem Biol, 17 (11): 1241-9. [PMID:21095574]
How to cite this page
p38 subfamily: mitogen-activated protein kinase 12. Last modified on 09/08/2022. Accessed on 14/12/2025. IUPHAR/BPS Guide to PHARMACOLOGY, https://www.guidetoimmunopharmacology.org/GRAC/ObjectDisplayForward?objectId=1501.