|
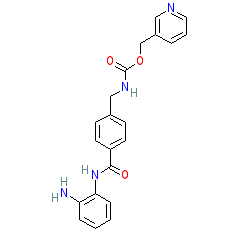
Synonyms: BAY 86-5274 | MS-275 | SNDX-275 | ZK 244894
Compound class:
Synthetic organic
Comment: Entinostat (MS-275) inhibits Class I histone deacetylases (HDACs 1, 2, 3 & 8), but has been shown to be somewhat selective for HDAC1 [1,4]. Studies in rodent models of inflammation suggest that inhibition by entinostat may be clinically useful [2,6-8], perhaps more so than broader acting Class I HDAC inhibitors such as vorinostat (SAHA) [6].
Ligand Activity Visualisation ChartsThese are box plot that provide a unique visualisation, summarising all the activity data for a ligand taken from ChEMBL and GtoPdb across multiple targets and species. Click on a plot to see the median, interquartile range, low and high data points. A value of zero indicates that no data are available. A separate chart is created for each target, and where possible the algorithm tries to merge ChEMBL and GtoPdb targets by matching them on name and UniProt accession, for each available species. However, please note that inconsistency in naming of targets may lead to data for the same target being reported across multiple charts. ✖ |
|
|||||||||||||||||||||||||||||||||||
| Selectivity at enzymes | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||