GtoPdb is requesting financial support from commercial users. Please see our sustainability page for more information.
SET and MYND domain containing 2
Target not currently curated in GtoImmuPdb
Target id: 2714
Nomenclature: SET and MYND domain containing 2
Contents:
Gene and Protein Information  |
||||||
| Species | TM | AA | Chromosomal Location | Gene Symbol | Gene Name | Reference |
| Human | - | 433 | 1q32.3 | SMYD2 | SET and MYND domain containing 2 | |
| Mouse | - | 433 | 1 H6 | Smyd2 | SET and MYND domain containing 2 | |
| Rat | - | 433 | 13q27 | Smyd2 | SET and MYND domain containing 2 | |
Previous and Unofficial Names  |
| KMT3C |
Database Links  |
|
| Alphafold | Q9NRG4 (Hs), Q8R5A0 (Mm), Q7M6Z3 (Rn) |
| BRENDA | 2.1.1.43 |
| CATH/Gene3D | 1.25.40.10 |
| ChEMBL Target | CHEMBL2169716 (Hs) |
| Ensembl Gene | ENSG00000143499 (Hs), ENSMUSG00000026603 (Mm), ENSRNOG00000003583 (Rn) |
| Entrez Gene | 56950 (Hs), 226830 (Mm), 289372 (Rn) |
| Human Protein Atlas | ENSG00000143499 (Hs) |
| KEGG Enzyme | 2.1.1.43 |
| KEGG Gene | hsa:56950 (Hs), mmu:226830 (Mm), rno:289372 (Rn) |
| OMIM | 610663 (Hs) |
| Pharos | Q9NRG4 (Hs) |
| RefSeq Nucleotide | NM_020197 (Hs), NM_026796 (Mm), NM_206851 (Rn) |
| RefSeq Protein | NP_064582 (Hs), NP_081072 (Mm), NP_996733 (Rn) |
| SynPHARM |
79343 (in complex with AZ505) 83226 (in complex with BAY-598) 81854 (in complex with LLY-507) |
| UniProtKB | Q9NRG4 (Hs), Q8R5A0 (Mm), Q7M6Z3 (Rn) |
| Wikipedia | SMYD2 (Hs) |
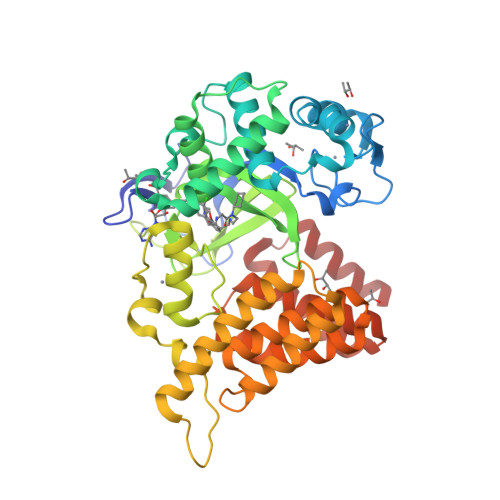
Selected 3D Structures  |
|||||||||||||
|
|
||||||||||||
Enzyme Reaction  |
||||
|
||||
Download all structure-activity data for this target as a CSV file 
| Inhibitors | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | View all chemical structures | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| General Comments |
| SMYD2 methylates histone and non-histone proteins. The enzyme methylates histone H3 ' at lusine 4 (H3K4me) and dimethylates histone H3 at lysine 36 (H3K36me2) [1]. SMYD2 modulation of chromatin structure is reported to modulate cell proliferation [1]. |
References
1. Brown MA, Sims 3rd RJ, Gottlieb PD, Tucker PW. (2006) Identification and characterization of Smyd2: a split SET/MYND domain-containing histone H3 lysine 36-specific methyltransferase that interacts with the Sin3 histone deacetylase complex. Mol Cancer, 5: 26. [PMID:16805913]
2. Ferguson AD, Larsen NA, Howard T, Pollard H, Green I, Grande C, Cheung T, Garcia-Arenas R, Cowen S, Wu J et al.. (2011) Structural basis of substrate methylation and inhibition of SMYD2. Structure, 19 (9): 1262-73. [PMID:21782458]
3. SGC. BAY-598 A selective chemical probe for SMYD2. Accessed on 11/12/2015. Modified on 04/08/2023. thesgc.org, https://www.thesgc.org/chemical-probes/BAY-598
4. SGC. LLY-507: A chemical probe for SMYD2 protein lysine methyltransferase. Accessed on 03/03/2015. Modified on 04/08/2023. thesgc.org, https://www.thesgc.org/chemical-probes/LLY-507
How to cite this page
2.1.1.43 Histone methyltransferases (HMTs): SET and MYND domain containing 2. Last modified on 11/12/2015. Accessed on 31/01/2026. IUPHAR/BPS Guide to PHARMACOLOGY, https://www.guidetoimmunopharmacology.org/GRAC/ObjectDisplayForward?objectId=2714.