GtoPdb is requesting financial support from commercial users. Please see our sustainability page for more information.
|
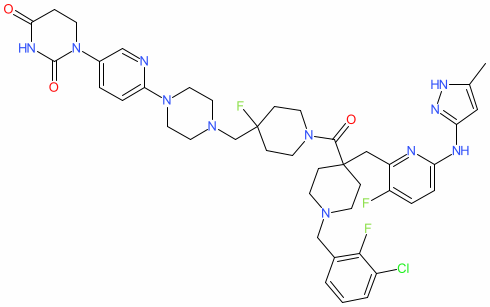
Synonyms: compound 33 [PMID: 41252673] | SK-5527
Compound class:
Synthetic organic
Comment: SK5527 is an Aurora kinase A (AURKA) PROTAC degrader [1]. SK5527 was developed as a progression from the previously reported MK-5108-thalidomide cereblon E3-recruiting AURKA PROTAC SK2188 [2], which was reported to induce loss of the neuroblastoma-related oncogenic transcription factor MYCN (MYCN), but whose pharmacokinetics precluded in vivo activity; AURKA normally protects MYCN from proteasomal degradation by the SCFFBXW7 E3 ubiquitin ligase.
Ligand Activity Visualisation ChartsThese are box plot that provide a unique visualisation, summarising all the activity data for a ligand taken from ChEMBL and GtoPdb across multiple targets and species. Click on a plot to see the median, interquartile range, low and high data points. A value of zero indicates that no data are available. A separate chart is created for each target, and where possible the algorithm tries to merge ChEMBL and GtoPdb targets by matching them on name and UniProt accession, for each available species. However, please note that inconsistency in naming of targets may lead to data for the same target being reported across multiple charts. ✖ |
|
|||||||||||||||||||||||||||||||||||
| Bioactivity Comments |
| SK5527 is >5000-fold selective for AURKA over AURKB in terms of binding affinity. |
| Selectivity at enzymes | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||