GtoPdb is requesting financial support from commercial users. Please see our sustainability page for more information.
|
Compound class:
Synthetic organic
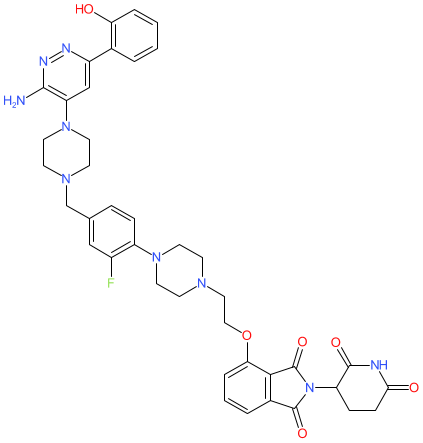
Comment: YD54 is an orally bioavailable, cereblon-engaging PROTAC degrader of SWI/SNF related BAF chromatin remodeling complex subunit ATPase 2 (SMARCA2) [3]. It is a bifunctional molecule containing a SMARCA2 bromodomain binding ligand linked to pomalidomide (to recruit the cereblon E3 ubiquitin ligase). Disrupting SMARCA2 is synthetic lethal in lung cancer cells that harbour loss of function mutations in the paralog SMARCA4 [1-2,4-5].
|
|
|||||||||||||||||||||||||||||||||||
| References |
|
1. Farnaby W, Koegl M, Roy MJ, Whitworth C, Diers E, Trainor N, Zollman D, Steurer S, Karolyi-Oezguer J, Riedmueller C et al.. (2019)
BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design. Nat Chem Biol, 15 (7): 672-680. [PMID:31178587] |
|
2. Hoffman GR, Rahal R, Buxton F, Xiang K, McAllister G, Frias E, Bagdasarian L, Huber J, Lindeman A, Chen D et al.. (2014)
Functional epigenetics approach identifies BRM/SMARCA2 as a critical synthetic lethal target in BRG1-deficient cancers. Proc Natl Acad Sci U S A, 111 (8): 3128-33. [PMID:24520176] |
|
3. Kotagiri S, Wang Y, Han Y, Liang X, Blazanin N, Mazhar H, Sebastian M, Nguyen PK, Jiang Y, Lissanu Y. (2025)
Discovery of Novel, Potent, and Orally Bioavailable SMARCA2 Proteolysis-Targeting Chimeras with Synergistic Antitumor Activity in Combination with Kirsten Rat Sarcoma Viral Oncogene Homologue G12C Inhibitors. J Med Chem, 68 (9): 9202-9219. [PMID:40280558] |
|
4. Oike T, Ogiwara H, Tominaga Y, Ito K, Ando O, Tsuta K, Mizukami T, Shimada Y, Isomura H, Komachi M et al.. (2013)
A synthetic lethality-based strategy to treat cancers harboring a genetic deficiency in the chromatin remodeling factor BRG1. Cancer Res, 73 (17): 5508-18. [PMID:23872584] |
|
5. Rago F, DiMare MT, Elliott G, Ruddy DA, Sovath S, Kerr G, Bhang HC, Jagani Z. (2019)
Degron mediated BRM/SMARCA2 depletion uncovers novel combination partners for treatment of BRG1/SMARCA4-mutant cancers. Biochem Biophys Res Commun, 508 (1): 109-116. [PMID:30527810] |