GtoPdb is requesting financial support from commercial users. Please see our sustainability page for more information.
Contents:
Gene and Protein Information  |
||||||
| Species | TM | AA | Chromosomal Location | Gene Symbol | Gene Name | Reference |
| SARS-CoV-2 | - | 306 | ||||
| SARS-CoV | - | 306 | ||||
| Gene and Protein Information Comments | ||||||
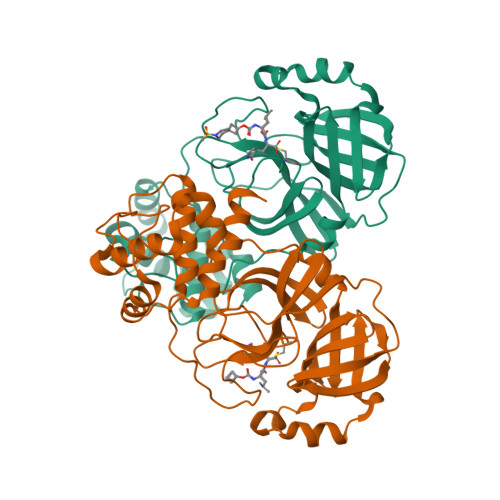
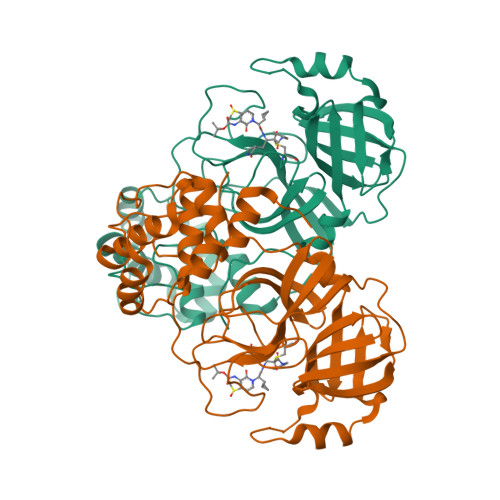
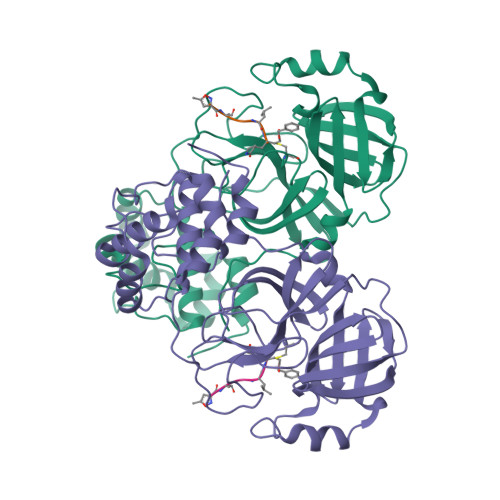
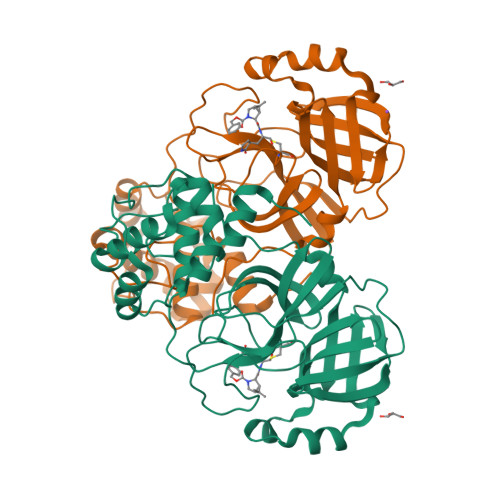
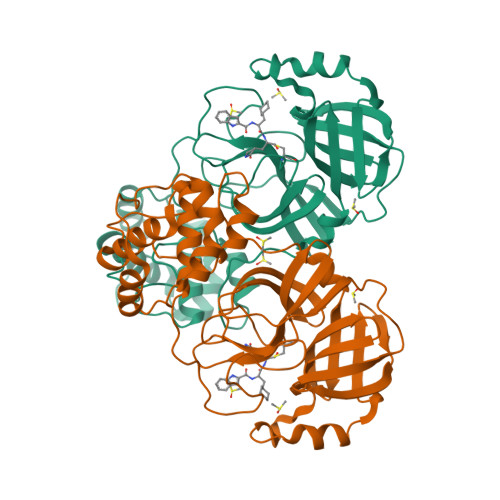
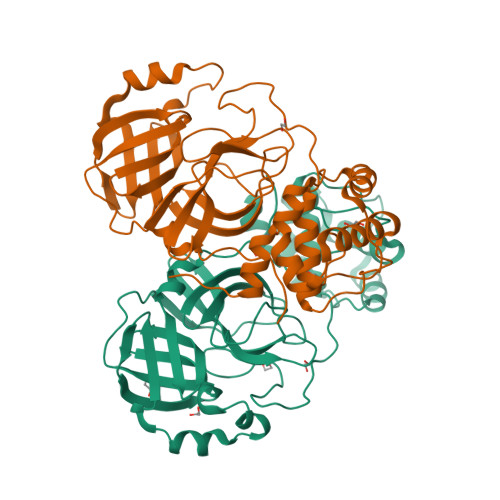
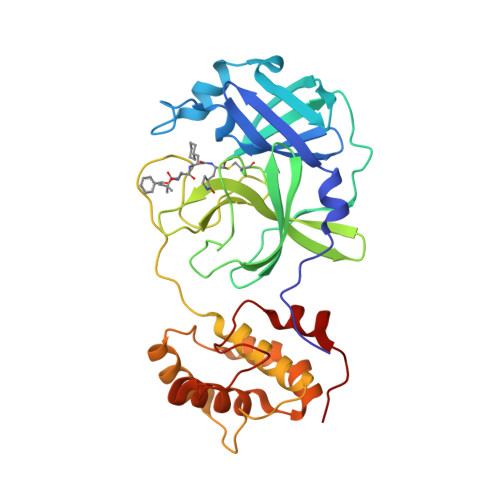
| The SARS-CoV main protease (Mpro) is a 306 amino acid cysteine protease that is encoded in the viral RNA replicase gene. It is amino acids 3241-3546 of the full length SARS-CoV polyprotein (3919 amino acids). The SARS-CoV-2 Mpro is also 306 amino acids (3264-3569 of the full length polyprotein). The RefSeq YP_009725301 was allocated to SARS-CoV-2 Mpro. The protein has been crystalised in complex with the inhibitor PRD_002214 (N3), and the structure was submitted to the RCSB Protein Databank with ID 6LU7 [50]. The UniProt ID P0DTD1 refers to the complete SARS-CoV-2 replicase polyprotein (length 7096 amino acids). Mpro has been assigned the IUBMB enzyme nomenclature identifier EC 3.4.22.69. |
||||||
Previous and Unofficial Names  |
| 3c-like proteinase | SARS-CoV-2 Mpro | Chain A, 3c-like Proteinase | 3CL protease | Mpro | nsp5 |
Database Links  |
|
| ChEMBL Target | CHEMBL3927 (SARS-CoV) |
| RefSeq Protein | YP_009725301 (SARS-CoV-2) |
| UniProtKB | P0C6U8 (SARS-CoV) |
Selected 3D Structures  |
|||||||||||||
|
|
||||||||||||
|
|
||||||||||||
|
|
||||||||||||
|
|
||||||||||||
|
|
||||||||||||
|
|
||||||||||||
|
|
||||||||||||
|
|
||||||||||||
|
|
||||||||||||
|
|
||||||||||||
|
|
||||||||||||
|
|
||||||||||||
|
|
||||||||||||
|
|
||||||||||||
|
|
||||||||||||
|
|
||||||||||||

|
|
||||||||||||
|
|
||||||||||||
| EC number (SARS-CoV-2) |
| 3.4.22.69 |
Download all structure-activity data for this target as a CSV file 
| Inhibitors | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | View all chemical structures | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| View species-specific inhibitor tables | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Inhibitor Comments | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PRD_002214 (N3) inhibits SARS-CoV-2 plaque formation in Vero cell culture with an IC50 of 16.77 μM [50]. The MERS-CoV inhibitor compound 11r [PMID: 32045235] is active against SARS-CoV-2 [108]. Baker et al. (2021) performed a large scale repurposing screen, which corroborated the inhibitory activity of boceprevir (and other hepatitis C NS3/4A protease inhibitors) against SARS-CoV-2 Mpro [5]. Chia et al. published a review of patents claiming coronavirus 3CLpro inhibitors in October 2021 [17]. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Other Binding Ligands | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | Click column headers to sort | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| General Comments |
|
The coronavirus (CoV) main proteases (Mpro) are cysteine proteases that are encoded in the viral RNA replicase gene. Mpro catalyses the proteolytic processing (cleavage) of replicase precursor polyproteins in to discrete functional proteins. In total There are 11 Mpro cleavage site within the C-terminus of the replicase polyprotein. Mpro plays a central role in the viral life cycle, and in light of evidence from other coronaviruses, SARS-CoV Mpro was a lead target for antiviral drug discovery. Many of the compounds that were discovered to inhibit the activity of MERS- and SARS-CoV Mpro enzymes have been tested for activity against SARS-CoV-2 [77]. Inhibitor design and development in response to SARS-CoV-2 has been intense [53]. Mpro has been reported to induce damage to the microvascular network in the brain, via proteolytic cleavage and inactivation of the endothelially-expressed protein NEMO (an essential NF-κB modulator with a central role in immunity; HGNC symbol IKBKG) [96]. Depletion of NEMO leads to cell death, and this cell death is dependent on receptor-interacting protein kinase 3 (RIPK3) signalling. A small molecule inhibitor of RIPK1, an upstream kinase that activates RIPK3, was shown to block the Mpro-induced microvascular pathology. It is interesting to note that a RIPK1 inhibitor (SAR443122) is already under clinical evaluation in COVID-19 patients. Although Mpro is strictly a component of the CoV replicase polyprotein(s) 1a and 1ab, we have included it as a separate entity to allow us to more sensibly curate pharmacological information (particularly regarding inhibitor development) that is specific for this protease, and to facilitate data retrieval. |
References
1. Alugubelli YR, Xiao J, Khatua K, Kumar S, Sun L, Ma Y, Ma XR, Vulupala VR, Atla S, Blankenship LR et al.. (2024) Discovery of First-in-Class PROTAC Degraders of SARS-CoV-2 Main Protease. J Med Chem, 67 (8): 6495-6507. [PMID:38608245]
2. Anson BJ, Chapman ME, Lendy EK, Pshenychnyi S, D’Aquila RT, Satchell KJF, Mesecar AD. (2020) Broad-spectrum inhibition of coronavirus main and papain-like proteases by HCV drugs. Nature Research, PrePrint, Under Review. DOI: 10.21203/rs.3.rs-26344/v1
3. Arnold LD, Jennings A, Keung W. (2021) Inhibitors of cysteine proteases and methods of use thereof. Patent number: WO2021252644A1. Assignee: Pardes Biosciences, Inc.. Priority date: 09/06/2020. Publication date: 16/12/2021.
4. Bai B, Belovodskiy A, Hena M, Kandadai AS, Joyce MA, Saffran HA, Shields JA, Khan MB, Arutyunova E, Lu J et al.. (2022) Peptidomimetic α-Acyloxymethylketone Warheads with Six-Membered Lactam P1 Glutamine Mimic: SARS-CoV-2 3CL Protease Inhibition, Coronavirus Antiviral Activity, and in Vitro Biological Stability. J Med Chem, 65 (4): 2905-2925. [PMID:34242027]
5. Baker JD, Uhrich RL, Kraemer GC, Love JE, Kraemer BC. (2021) A drug repurposing screen identifies hepatitis C antivirals as inhibitors of the SARS-CoV2 main protease. PLoS One, 16 (2): e0245962. [PMID:33524017]
6. Bao D, Guo F, Hesse MJ, Hornak V, Joseph S, Kirrane TM Jr., Liu B, Lin H, Miao Y, Moser HE et al.. (2023) Compounds and compositions for the treatment of coronaviral related diseases. Patent number: WO2023002409A1. Assignee: Novartis AG. Priority date: 20/07/2022. Publication date: 26/01/2023.
7. Boby ML, Fearon D, Ferla M, Filep M, Koekemoer L, Robinson MC, COVID Moonshot Consortium‡, Chodera JD, Lee AA, London N et al.. (2023) Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors. Science, 382 (6671): eabo7201. [PMID:37943932]
8. Botyanszki J, Catalano G, Chong PY, Dickson H, Jin Q, Leivers A, Maynard A, Liao X, Miller J, Shotwell JB et al.. (2018) Compounds that inhibit 3c and 3cl proteases and methods of use thereof. Patent number: WO2018042343A2. Assignee: Glaxosmithkline Intellectual Property (No.2) Limited. Priority date: 30/08/2016. Publication date: 08/03/2018.
9. Breidenbach J, Lemke C, Pillaiyar T, Schäkel L, Al Hamwi G, Diett M, Gedschold R, Geiger N, Lopez V, Mirza S et al.. (2021) Targeting the Main Protease of SARS-CoV-2: From the Establishment of High Throughput Screening to the Design of Tailored Inhibitors. Angew Chem Int Ed Engl, 60 (18): 10423-10429. [PMID:33655614]
10. Breidenbach J, Voget R, Si Y, Hingst A, Claff T, Sylvester K, Wolf V, Krasniqi V, Useini A, Sträter N et al.. (2024) Macrocyclic Azapeptide Nitriles: Structure-Based Discovery of Potent SARS-CoV-2 Main Protease Inhibitors as Antiviral Drugs. J Med Chem, 67 (11): 8757-8790. [PMID:38753594]
11. Carlsson J, Danielson H, Moodie L, Sandstrom A. (2022) SUBSTITUTED HYDANTOIN COMPOUNDS, METHODS FOR PREPARATION THEREOF AND USE THEREOF IN THE TREATMENT AND/OR PREVENTION OF A CORONA VIRUS DISEASE. Patent number: WO2022229458. Priority date: 22/06/2021. Publication date: 03/11/2022.
12. Chakraborty S, Mallick D, Goswami M, Guengerich FP, Chakrabarty A, Chowdhury G. (2022) The Natural Products Withaferin A and Withanone from the Medicinal Herb Withania somnifera Are Covalent Inhibitors of the SARS-CoV-2 Main Protease. J Nat Prod, 85 (10): 2340-2350. [PMID:36098617]
13. Chamakuri S, Lu S, Ucisik MN, Bohren KM, Chen YC, Du HC, Faver JC, Jimmidi R, Li F, Li JY et al.. (2021) DNA-encoded chemistry technology yields expedient access to SARS-CoV-2 Mpro inhibitors. Proc Natl Acad Sci U S A, 118 (36). DOI: 10.1073/pnas.2111172118 [PMID:34426525]
14. Chen J, Liang C, Miao K, Wu Y, Yun H, Zhang W. (2022) Aminocarbamoyl compounds for the treatment of viral infections. Patent number: WO2022043374. Assignee: Hoffman La Roche. Priority date: 25/08/2021. Publication date: 03/03/2022.
15. Chen X, Huang X, Ma Q, Kuzmič P, Zhou B, Zhang S, Chen J, Xu J, Liu B, Jiang H et al.. (2024) Preclinical evaluation of the SARS-CoV-2 Mpro inhibitor RAY1216 shows improved pharmacokinetics compared with nirmatrelvir. Nat Microbiol, 9 (4): 1075-1088. [PMID:38553607]
16. Chia CSB, Tan QW, Vankadara S, See YY, Ng Sp, Xu W, Lpw CH. (2022) Coronavirus enzyme modulators, methods of synthesis and uses thereof. Patent number: WO2022265577A2. Assignee: Agency For Science, Technology And Research. Priority date: 14/06/2022. Publication date: 22/12/2022.
17. Chia CSB, Xu W, Shuyi Ng P. (2022) A Patent Review on SARS Coronavirus Main Protease (3CLpro ) Inhibitors. ChemMedChem, 17 (1): e202100576. [PMID:34651447]
18. Chuck CP, Chen C, Ke Z, Wan DC, Chow HF, Wong KB. (2013) Design, synthesis and crystallographic analysis of nitrile-based broad-spectrum peptidomimetic inhibitors for coronavirus 3C-like proteases. Eur J Med Chem, 59: 1-6. [PMID:23202846]
19. Congreve MS, Christopher JA, Pickworth M, De Graff C, Higueruelo AP, Mason JS, Kulkarni SS. (2022) SARS-COV-2 MPRO INHIBITOR COMPOUNDS. Patent number: WO2022129953. Assignee: HEPTARES THERAPEUTICS LIMITED. Priority date: 18/12/2021. Publication date: 23/06/2022.
20. Cooper MS, Zhang L, Ibrahim M, Zhang K, Sun X, Röske J, Göhl M, Brönstrup M, Cowell JK, Sauerhering L et al.. (2022) Diastereomeric Resolution Yields Highly Potent Inhibitor of SARS-CoV-2 Main Protease. J Med Chem, 65 (19): 13328-13342. [PMID:36179320]
21. Cui J, Jia J. (2021) Discovery of juglone and its derivatives as potent SARS-CoV-2 main proteinase inhibitors. Eur J Med Chem, 225: 113789. [PMID:34438124]
22. Dai W, Jochmans D, Xie H, Yang H, Li J, Su H, Chang D, Wang J, Peng J, Zhu L et al.. (2022) Design, Synthesis, and Biological Evaluation of Peptidomimetic Aldehydes as Broad-Spectrum Inhibitors against Enterovirus and SARS-CoV-2. J Med Chem, 65 (4): 2794-2808. [PMID:33872498]
23. Dai W, Zhang B, Jiang XM, Su H, Li J, Zhao Y, Xie X, Jin Z, Peng J, Liu F et al.. (2020) Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science, 368 (6497): 1331-1335. [PMID:32321856]
24. Dampalla CS, Kim Y, Bickmeier N, Rathnayake AD, Nguyen HN, Zheng J, Kashipathy MM, Baird MA, Battaile KP, Lovell S et al.. (2021) Structure-Guided Design of Conformationally Constrained Cyclohexane Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3CL Protease. J Med Chem, 64 (14): 10047-10058. [PMID:34213885]
25. Dampalla CS, Kim Y, Zabiegala A, Howard DJ, Nguyen HN, Madden TK, Thurman HA, Cooper A, Liu L, Battaile KP et al.. (2024) Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies. J Med Chem, 67 (14): 11937-11956. [PMID:38953866]
26. Dampalla CS, Rathnayake AD, Galasiti Kankanamalage AC, Kim Y, Perera KD, Nguyen HN, Miller MJ, Madden TK, Picard HR, Thurman HA et al.. (2022) Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease. Journal of Medicinal Chemistry,. DOI: 10.1021/acs.jmedchem.2c00224
27. Dampalla CS, Rathnayake AD, Perera KD, Jesri AM, Nguyen HN, Miller MJ, Thurman HA, Zheng J, Kashipathy MM, Battaile KP et al.. (2021) Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies. J Med Chem, 64 (24): 17846-17865. [PMID:34865476]
28. Dampalla CS, Zheng J, Perera KD, Wong LR, Meyerholz DK, Nguyen HN, Kashipathy MM, Battaile KP, Lovell S, Kim Y et al.. (2021) Postinfection treatment with a protease inhibitor increases survival of mice with a fatal SARS-CoV-2 infection. Proc Natl Acad Sci U S A, 118 (29). DOI: 10.1073/pnas.2101555118 [PMID:34210738]
29. Dayan Elshan NGR, Wolff KC, Riva L, Woods AK, Grabovyi G, Wilson K, Pedroarena J, Ghorai S, Nazarian A, Weiss F et al.. (2024) Discovery of CMX990: A Potent SARS-CoV-2 3CL Protease Inhibitor Bearing a Novel Warhead. J Med Chem, 67 (4): 2369-2378. [PMID:38335279]
30. Drayman R, DeMarco JK, Jones KA, Azizi S-A, Froggatt HM, Tan K, Maltseva NI, Chen S, Nicolaescu V, Dvorkin S et al.. (2021) Masitinib is a broad coronavirus 3CL inhibitor that blocks replication of SARS-CoV-2. Science, {Epub ahead of print]. DOI: 10.1126/science.abg5827
31. Elseginy SA, Fayed B, Hamdy R, Mahrous N, Mostafa A, Almehdi AM, S M Soliman S. (2021) Promising anti-SARS-CoV-2 drugs by effective dual targeting against the viral and host proteases. Bioorg Med Chem Lett, 43: 128099. [PMID:33984473]
32. Fàbrega-Ferrer M, Herrera-Morandé A, Muriel-Goñi S, Pérez-Saavedra J, Bueno P, Castro V, Garaigorta U, Gastaminza P, Coll M. (2022) Structure and inhibition of SARS-CoV-1 and SARS-CoV-2 main proteases by oral antiviral compound AG7404. Antiviral Res, 208: 105458. [PMID:36336176]
33. Flury P, Krüger N, Sylvester K, Breidenbach J, Al Hamwi G, Qiao J, Chen Y, Rocha C, Serafim MSM, Barbosa da Silva E et al.. (2025) Design, Synthesis, and Unprecedented Interactions of Covalent Dipeptide-Based Inhibitors of SARS-CoV-2 Main Protease and Its Variants Displaying Potent Antiviral Activity. J Med Chem, 68 (3): 3626-3652. [PMID:39813204]
34. Geng ZZ, Atla S, Shaabani N, Vulupala V, Yang KS, Alugubelli YR, Khatua K, Chen PH, Xiao J, Blankenship LR et al.. (2023) A Systematic Survey of Reversibly Covalent Dipeptidyl Inhibitors of the SARS-CoV-2 Main Protease. J Med Chem, 66 (16): 11040-11055. [PMID:37561993]
35. Ghosh AK, Gong G, Grum-Tokars V, Mulhearn DC, Baker SC, Coughlin M, Prabhakar BS, Sleeman K, Johnson ME, Mesecar AD. (2008) Design, synthesis and antiviral efficacy of a series of potent chloropyridyl ester-derived SARS-CoV 3CLpro inhibitors. Bioorg Med Chem Lett, 18 (20): 5684-8. [PMID:18796354]
36. Ghosh AK, Raghavaiah J, Shahabi D, Yadav M, Anson BJ, Lendy EK, Hattori SI, Higashi-Kuwata N, Mitsuya H, Mesecar AD. (2021) Indole Chloropyridinyl Ester-Derived SARS-CoV-2 3CLpro Inhibitors: Enzyme Inhibition, Antiviral Efficacy, Structure-Activity Relationship, and X-ray Structural Studies. J Med Chem, 64 (19): 14702-14714. [PMID:34528437]
37. Glaser J, Sedova A, Galanie S, Kneller DW, Davidson RB, Maradzike E, Del Galdo S, Labbé A, Hsu DJ, Agarwal R et al.. (2022) Hit Expansion of a Noncovalent SARS-CoV-2 Main Protease Inhibitor. ACS Pharmacol Transl Sci, 5 (4): 255-265. [PMID:35434531]
38. Grifagni D, Lenci E, De Santis A, Orsetti A, Barracchia CG, Tedesco F, Bellini Puglielli R, Lucarelli F, Lauriola A, Assfalg M et al.. (2024) Development of a GC-376 Based Peptidomimetic PROTAC as a Degrader of 3-Chymotrypsin-like Protease of SARS-CoV-2. ACS Med Chem Lett, 15 (2): 250-257. [PMID:38352832]
39. Griffen EJ, Cousins DL, Stacey-Jepson JE, Lefker BA, Robinson RP Jr. (2025) CHEMICAL COMPOUNDSID50000167790940 P. DOC 2025-05-07 FILING NO.:19. Patent number: WO2025125695. Assignee: MEDCHEMICA CONSULTANCY LTD, DRUGS FOR NEGLECTED DISEASES INITIATIVE. Priority date: 17/12/2024. Publication date: 19/06/2025.
40. Griffen EJ, Fearon D, McGovern BL, Koekemoer L, Balcomb BH, Szommer T, Fate G, Robinson RP, Lefker BA, Duberstein S et al.. (2025) Open-science discovery of DNDI-6510, a compound that addresses genotoxic and metabolic liabilities of the COVID Moonshot SARS-CoV-2 Mpro lead inhibitor. , Preprint. DOI: 10.1101/2025.06.16.660018
41. Hamdy R, Fayed B, Mostafa A, Shama NMA, Mahmoud SH, Mehta CH, Nayak Y, M Soliman SS. (2021) Iterated Virtual Screening-Assisted Antiviral and Enzyme Inhibition Assays Reveal the Discovery of Novel Promising Anti-SARS-CoV-2 with Dual Activity. Int J Mol Sci, 22 (16). [PMID:34445763]
42. Han SH, Goins CM, Arya T, Shin WJ, Maw J, Hooper A, Sonawane DP, Porter MR, Bannister BE, Crouch RD et al.. (2022) Structure-Based Optimization of ML300-Derived, Noncovalent Inhibitors Targeting the Severe Acute Respiratory Syndrome Coronavirus 3CL Protease (SARS-CoV-2 3CLpro). J Med Chem, 65 (4): 2880-2904. [PMID:34347470]
43. Hattori SI, Higashi-Kuwata N, Hayashi H, Allu SR, Raghavaiah J, Bulut H, Das D, Anson BJ, Lendy EK, Takamatsu Y et al.. (2021) A small molecule compound with an indole moiety inhibits the main protease of SARS-CoV-2 and blocks virus replication. Nat Commun, 12 (1): 668. [PMID:33510133]
44. Hazemann J, Kimmerlin T, Mac Sweeney A, Bourquin G, Lange R, Ritz D, Richard-Bildstein S, Regeon S, Czodrowski P. (2025) Accelerating the Hit-To-Lead Optimization of a SARS-CoV-2 Mpro Inhibitor Series by Combining High-Throughput Medicinal Chemistry and Computational Simulations. J Med Chem, [Epub ahead of print]. [PMID:40186586]
45. Hirose Y, Shindo N, Mori M, Onitsuka S, Isogai H, Hamada R, Hiramoto T, Ochi J, Takahashi D, Ueda T et al.. (2022) Discovery of Chlorofluoroacetamide-Based Covalent Inhibitors for Severe Acute Respiratory Syndrome Coronavirus 2 3CL Protease. J Med Chem, 65 (20): 13852-13865. [PMID:36229406]
46. Hoffman RL, Kania RS, Brothers MA, Davies JF, Ferre RA, Gajiwala KS, He M, Hogan RJ, Kozminski K, Li LY et al.. (2020) Discovery of Ketone-Based Covalent Inhibitors of Coronavirus 3CL Proteases for the Potential Therapeutic Treatment of COVID-19. J Med Chem, 63 (21): 12725-12747. [PMID:33054210]
47. Hou N, Shuai L, Zhang L, Xie X, Tang K, Zhu Y, Yu Y, Zhang W, Tan Q, Zhong G et al.. (2023) Development of Highly Potent Noncovalent Inhibitors of SARS-CoV-2 3CLpro. ACS Cent Sci, 9 (2): 217-227. [PMID:36844503]
48. Hu S, Zhang Y, Wang C, Li J, Su H, Xie X, Wang J, Wang J, Cao J, He X et al.. (2025) Development of Orally Bioavailable Octahydroindole-Based Peptidomimetic Derivative as a Broad-Spectrum Inhibitor against HCoV-OC43 and SARS-CoV-2. J Med Chem, 68 (11): 10823-10844. [PMID:40400488]
49. Huang C, Shuai H, Qiao J, Hou Y, Zeng R, Xia A, Xie L, Fang Z, Li Y, Yoon C et al.. (2023) A new generation Mpro inhibitor with potent activity against SARS-CoV-2 Omicron variants. Signal Transduct Target Ther, 8 (1): 128. [PMID:36928316]
50. Jin Z, Du X, Xu Y, Deng Y, Liu M, Zhang B, Li X, Zhang L, Peng C, Duan Y. (2020) Structure of Mpro from COVID-19 virus and discovery of its inhibitors. bioRxiv, Preprint. DOI: 10.1101/2020.02.26.964882
51. Konno S, Kobayashi K, Senda M, Funai Y, Seki Y, Tamai I, Schäkel L, Sakata K, Pillaiyar T, Taguchi A et al.. (2022) 3CL Protease Inhibitors with an Electrophilic Arylketone Moiety as Anti-SARS-CoV-2 Agents. J Med Chem, 65 (4): 2926-2939. [PMID:34313428]
52. Konno S, Thanigaimalai P, Yamamoto T, Nakada K, Kakiuchi R, Takayama K, Yamazaki Y, Yakushiji F, Akaji K, Kiso Y et al.. (2013) Design and synthesis of new tripeptide-type SARS-CoV 3CL protease inhibitors containing an electrophilic arylketone moiety. Bioorg Med Chem, 21 (2): 412-24. [PMID:23245752]
53. La Monica G, Bono A, Lauria A, Martorana A. (2022) Targeting SARS-CoV-2 Main Protease for Treatment of COVID-19: Covalent Inhibitors Structure-Activity Relationship Insights and Evolution Perspectives. J Med Chem, 65 (19): 12500-12534. [PMID:36169610]
54. Lee CC, Kuo CJ, Ko TP, Hsu MF, Tsui YC, Chang SC, Yang S, Chen SJ, Chen HC, Hsu MC et al.. (2009) Structural basis of inhibition specificities of 3C and 3C-like proteases by zinc-coordinating and peptidomimetic compounds. J Biol Chem, 284 (12): 7646-55. [PMID:19144641]
55. Liang C, Xin L, Tian L, Xia J, Qin N, Li J, Qiang T, Li H, Wang X, Xie X et al.. (2022) Protacs based on VHL ligand targeting coronavirus 3CL protease and preparation method and application thereof. Patent number: US11518759B1. Assignee: Shaanxi Panlong Pharmaceutical Co Ltd. Priority date: 23/06/2022. Publication date: 06/12/2022.
56. Liang C, Xin L, Tian L, Xia J, Qin N, Li J, Qiang T, Li H, Wang X, Xie X et al.. (2022) Protacs targeting coronavirus 3CL protease and preparation method and application thereof. Patent number: US11530195B1. Assignee: Shaanxi Panlong Pharmaceutical Co Ltd. Priority date: 20/05/2022. Publication date: 20/12/2022.
57. Liu H, Iketani S, Zask A, Khanizeman N, Bednarova E, Forouhar F, Fowler B, Hong SJ, Mohri H, Nair MS et al.. (2022) Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19. Nat Commun, 13 (1): 1891. [PMID:35393402]
58. Liu R, Xu Y, Hua L, Zhou J, Deng H, Chu X, Ding S. (2023) 3CL PROTEASE SMALL-MOLECULE INHIBITOR FOR TREATING OR PREVENTING CORONAVIRUS INFECTION, AND USE THEREOF. Patent number: WO2023011443. Assignee: THE GLOBAL HEALTH DRUG DISCOVERY INSTITUTE [CN]. Priority date: 02/08/2021. Publication date: 09/02/2023.
59. Luttens A, Gullberg H, Abdurakhmanov E, Vo DD, Akaberi D, Talibov VO, Nekhotiaeva N, Vangeel L, De Jonghe S, Jochmans D et al.. (2022) Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses. J Am Chem Soc, 144 (7): 2905-2920. [PMID:35142215]
60. Ma C, Sacco MD, Hurst B, Townsend JA, Hu Y, Szeto T, Zhang X, Tarbet B, Marty MT, Chen Y et al.. (2020) Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res, 30 (8): 678-692. DOI: 10.1038/s41422-020-0356-z [PMID:32541865]
61. Mao L, Shaabani N, Zhang X, Jin C, Xu W, Argent C, Kushnareva Y, Powers C, Stegman K, Liu J et al.. (2024) Olgotrelvir, a dual inhibitor of SARS-CoV-2 Mpro and cathepsin L, as a standalone antiviral oral intervention candidate for COVID-19. Med, 5 (1): 42-61.e23. [PMID:38181791]
62. Matos-Hernández ML, Samples R, Dyer G, Casimir Montán VM, Morales-Colón CA, Salvino JM, Montaner LJ, Cassel JA, Messick TE, Tietjen I et al.. (2024) Metabolomic Analysis and Antiviral Screening of a Marine Algae Library Yield Jobosic Acid (2,5-Dimethyltetradecanoic Acid) as a Selective Inhibitor of SARS-CoV-2. J Nat Prod, 87 (6): 1513-1520. [PMID:38781491]
63. McGovern-Gooch KR, Mani N, Gotchev D, Ardzinski A, Kowalski R, Sheraz M, Micolochick Steuer HM, Tercero B, Wang X, Wasserman A et al.. (2024) Biological characterization of AB-343, a novel and potent SARS-CoV-2 Mpro inhibitor with pan-coronavirus activity. Antiviral Res, 232: 106038. [PMID:39577571]
64. Milligan JC, Zeisner TU, Papageorgiou G, Joshi D, Soudy C, Ulferts R, Wu M, Lim CT, Tan KW, Weissmann F et al.. (2021) Identifying SARS-CoV-2 antiviral compounds by screening for small molecule inhibitors of Nsp5 main protease. Biochem J, 478 (13): 2499-2515. [PMID:34198327]
65. Mitsuya H, Tamamura H, Kuwata N. (2023) COMPOUND EXHIBITING PHYSIOLOGICAL ACTIVITY SUCH AS ANTIVIRAL ACTIVITY. Patent number: WO2023286844. Assignee: NATIONAL CENTER FOR GLOBAL HEALTH AND MEDICINE [JP]. Priority date: 15/07/2021. Publication date: 10/01/2023.
66. Nieman JA, Lemieux MJ, Tyrrell DL, Bai B, Belovodskiy A, Hena M, Kandadai AS, Joyce MA. (2022) RNA VIRUS INHIBITOR COMPOUNDS AND USES THEREOF. Patent number: WO2022133588. Assignee: THE GOVERNORS OF THE UNIVERSITY OF ALBERTA [CA]. Priority date: 21/12/2020. Publication date: 30/06/2022.
67. Okabe A, Carney DW, Tawada M, Akther T, Aida J, Takagi T, Dougan DR, Leffler AE, Bell JA, Frye L et al.. (2025) Discovery of Highly Potent Noncovalent Inhibitors of SARS-CoV-2 Main Protease through Computer-Aided Drug Design. J Med Chem, 68 (20): 21330-21345. [PMID:41076627]
68. Owen DR, Allerton CMN, Anderson AS, Aschenbrenner L, Avery M, Berritt S, Boras B, Cardin RD, Carlo A, Coffman KJ et al.. (2021) An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19. Science, 374 (6575): 1586-1593. [PMID:34726479]
69. Owen DR, Pettersson MY, Reese MR, Sammona MF, Tuttle JB, Yang Q. (2022) Antiviral heteroaryl ketone derivatives. Patent number: WO2022013684A1. Assignee: Pfizer Inc.. Priority date: 07/07/2021. Publication date: 20/01/2022.
70. Owen DR, Reese MR, Sammons MF, Tuttle JB, Verhoest PR, Yang Q. (2022) Nitrile-Containing Antiviral Compounds. Patent number: US20220257563A1. Assignee: Pfizer Inc. Priority date: 22/04/2022. Publication date: 18/08/2022.
71. Pathak N, Chen YT, Hsu YC, Hsu NY, Kuo CJ, Tsai HP, Kang JJ, Huang CH, Chang SY, Chang YH et al.. (2021) Uncovering Flexible Active Site Conformations of SARS-CoV-2 3CL Proteases through Protease Pharmacophore Clusters and COVID-19 Drug Repurposing. ACS Nano, 15 (1): 857-872. [PMID:33373194]
72. Porzberg MRB, Groenewold GJM, Lyoo H, Jakob AKMH, Titulaer WHC, Cavina L, Poelaert KCK, Zwaagstra M, Dieteren CEJ, Lemmers JGH et al.. (2025) Peptidomimetic Phenoxymethyl Ketone Warheads as Potent Dual-Mode Inhibitors against SARS-CoV-2 Mpro and Cathepsin. J Med Chem, 68 (11): 10953-10969. [PMID:40415551]
73. PostEra AI. MPro Activity Data. Accessed on 11/08/2020. Modified on 11/08/2020. postera.ai/covid/activity_data, https://postera.ai/covid/activity_data
74. Previti S, Ettari R, Calcaterra E, Roggia M, Natale B, Weldert AC, Müller-Ruttloff C, Salisch F, Irto A, Cigala RM et al.. (2024) Identification of Dual Inhibitors Targeting Main Protease (Mpro) and Cathepsin L as Potential Anti-SARS-CoV-2 Agents. ACS Med Chem Lett, 15 (5): 602-609. [PMID:38746883]
75. Qiao J, Li YS, Zeng R, Liu FL, Luo RH, Huang C, Wang YF, Zhang J, Quan B, Shen C et al.. (2021) SARS-CoV-2 Mpro inhibitors with antiviral activity in a transgenic mouse model. Science, 371 (6536): 1374-1378. [PMID:33602867]
76. Rathnayake AD, Zheng J, Kim Y, Perera KD, Mackin S, Meyerholz DK, Kashipathy MM, Battaile KP, Lovell S, Perlman S et al.. (2020) 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice. Sci Transl Med, 12 (557). DOI: 10.1126/scitranslmed.abc5332 [PMID:32747425]
77. Riva L, Yuan S, Yin X, Martin-Sancho L, Matsunaga N, Pache L, Burgstaller-Muehlbacher S, De Jesus PD, Teriete P, Hull MV et al.. (2020) Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature, [Epub ahead of print]. DOI: 10.1038/s41586-020-2577-1
78. Rogovoy B, Kysil V, Berishvili V, Pauza CD, Zapata JC, Moreno SMM, Li A, Orry A, Lam PCH, Abagyan R. (2023) Compounds for treatment of a coronavirus infection. Patent number: WO2023245166A2. Assignee: Trawsfynydd Therapeutics. Priority date: 16/06/2023. Publication date: 21/12/2023.
79. Sá Magalhães Serafim M, Kronenberger T, Francisco KR, de Sousa Reis EV, Gonçalves de Oliveira E, Marcelino E Oliveira FK, Serraglio Fortes I, Maciel Fernandes TH, Barbosa da Silva E, Fajtova P et al.. (2025) Discovery of benzyl carbamate inhibitors of coronavirus Mpro enzymes from a legacy collection of cysteine protease inhibitors. J Enzyme Inhib Med Chem, 40 (1): 2585619. [PMID:41246830]
80. Sacco MD, Ma C, Lagarias P, Gao A, Townsend JA, Meng X, Dube P, Zhang X, Hu Y, Kitamura N et al.. (2020) Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against Mpro and cathepsin L. Sci Adv, 6 (50). [PMID:33158912]
81. Sato J, Shibayama H, Hirai K, Uno Y, Uehara S, Yonezawa S, Kurahashi K, Kojima E. (2023) Uracil derivative having viral growth inhibitory activity and pharmaceutical composition containing same. Patent number: WO2023195530A1. Assignee: Shionogi and Co Ltd. Priority date: 07/04/2023. Publication date: 12/10/2023.
82. Shen Z, Li Y, Zhu J, Huang Q, Yin J, Xu Y, Wu A, Su W, Kuai L. (2023) VIRUS MAIN PROTEASE INHIBITOR, PREPARATION METHOD THEREFOR, AND USE. Patent number: WO2023283831. Assignee: WUXI APPTEC (SHANGHAI) CO., LTD.. Priority date: 14/07/2021. Publication date: 19/01/2023.
83. Shurtleff VW, Layton ME, Parish CA, Perkins JJ, Schreier JD, Wang Y, Adam GC, Alvarez N, Bahmanjah S, Bahnck-Teets CM et al.. (2024) Invention of MK-7845, a SARS-CoV-2 3CL Protease Inhibitor Employing a Novel Difluorinated Glutamine Mimic. J Med Chem, 67 (5): 3935-3958. [PMID:38365209]
84. Tan B, Sacco M, Tan H, Li K, Joyce R, Zhang X, Chen Y, Wang J. (2023) Exploring diverse reactive warheads for the design of SARS-CoV-2 main protease inhibitors. Eur J Med Chem, 259: 115667. [PMID:37482021]
85. Taoda Y, Hori A, Tadano G, Sugiyama S, Masakado S, Tanaka S, Ogasahara R, Nakahara K, Maeno S, Unoh Y et al.. (2025) Design, synthesis and biological evaluation of novel quinazolinedione derivatives as non-covalent SARS-CoV-2 3CL protease inhibitors. Bioorg Med Chem Lett, 129: 130400 [Epub ahead of print]. [PMID:40939708]
86. Thanigaimalai P, Konno S, Yamamoto T, Koiwai Y, Taguchi A, Takayama K, Yakushiji F, Akaji K, Chen SE, Naser-Tavakolian A et al.. (2013) Development of potent dipeptide-type SARS-CoV 3CL protease inhibitors with novel P3 scaffolds: design, synthesis, biological evaluation, and docking studies. Eur J Med Chem, 68: 372-84. [PMID:23994330]
87. Thanigaimalai P, Konno S, Yamamoto T, Koiwai Y, Taguchi A, Takayama K, Yakushiji F, Akaji K, Kiso Y, Kawasaki Y et al.. (2013) Design, synthesis, and biological evaluation of novel dipeptide-type SARS-CoV 3CL protease inhibitors: structure-activity relationship study. Eur J Med Chem, 65: 436-47. [PMID:23747811]
88. The COVID Moonshot Consortium, Achdout H, Aimon A, Bar-David E, Barr H, Ben-Shmuel A, Bennett J, Boby ML, Borden B, Bowman GR et al.. (2021) Open Science Discovery of Oral Non-Covalent SARS-CoV-2 Main Protease Inhibitor Therapeutics. bioRxiv, Preprint. DOI: 10.1101/2020.10.29.339317
89. Tong X, Keung W, Arnold LD, Stevens LJ, Pruijssers AJ, Kook S, Lopatin U, Denison M, Kwong AD. (2023) Evaluation of in vitro antiviral activity of SARS-CoV-2 Mpro inhibitor pomotrelvir and cross-resistance to nirmatrelvir resistance substitutions. Antimicrob Agents Chemother, 67 (11): e0084023. [PMID:37800975]
90. Turlington M, Chun A, Tomar S, Eggler A, Grum-Tokars V, Jacobs J, Daniels JS, Dawson E, Saldanha A, Chase P et al.. (2013) Discovery of N-(benzo[1,2,3]triazol-1-yl)-N-(benzyl)acetamido)phenyl) carboxamides as severe acute respiratory syndrome coronavirus (SARS-CoV) 3CLpro inhibitors: identification of ML300 and noncovalent nanomolar inhibitors with an induced-fit binding. Bioorg Med Chem Lett, 23 (22): 6172-7. [PMID:24080461]
91. Unoh Y, Uehara S, Nakahara K, Nobori H, Yamatsu Y, Yamamoto S, Maruyama Y, Taoda Y, Kasamatsu K, Suto T et al.. (2022) Discovery of S-217622, a Noncovalent Oral SARS-CoV-2 3CL Protease Inhibitor Clinical Candidate for Treating COVID-19. J Med Chem, 65 (9): 6499-6512. [PMID:35352927]
92. Vandyck K, Raboisson P, Beigelman L, Serebryany V, Stoycheva A, Bardiot D, Boland S, Marchand A. (2021) Anti-viral compounds for treating coronavirus, picornavirus, and norovirus infections. Patent number: WO2021252491A1. Assignee: Aligos Therapeutics, Inc., Katholieke Universiteit Leuven. Priority date: 10/06/2020. Publication date: 16/12/2021.
93. Vankadara S, Wong YX, Liu B, See YY, Tan LH, Tan QW, Wang G, Karuna R, Guo X, Tan ST et al.. (2021) A head-to-head comparison of the inhibitory activities of 15 peptidomimetic SARS-CoV-2 3CLpro inhibitors. Bioorg Med Chem Lett, 48: 128263. [PMID:34271072]
94. Vuong W, Khan MB, Fischer C, Arutyunova E, Lamer T, Shields J, Saffran HA, McKay RT, van Belkum MJ, Joyce MA et al.. (2020) Feline coronavirus drug inhibits the main protease of SARS-CoV-2 and blocks virus replication. Nat Commun, 11 (1): 4282. [PMID:32855413]
95. Wang X, Gotchev D, Fan KY, Vega MM, Mani N, McGovern-Gooch K, Cuconati A, Tercero B, Wang X, Carpino P et al.. (2024) Rational Design of Macrocyclic Noncovalent Inhibitors of SARS-CoV-2 Mpro from a DNA-Encoded Chemical Library Screening Hit That Demonstrate Potent Inhibition against Pan-Coronavirus Homologues and Nirmatrelvir-Resistant Variants. J Med Chem, 67 (21): 19623-19667. [PMID:39453309]
96. Wenzel J, Lampe J, Müller-Fielitz H, Schuster R, Zille M, Müller K, Krohn M, Körbelin J, Zhang L, Özorhan Ü et al.. (2021) The SARS-CoV-2 main protease Mpro causes microvascular brain pathology by cleaving NEMO in brain endothelial cells. Nat Neurosci, 24 (11): 1522-1533. [PMID:34675436]
97. Westberg M, Su Y, Zou X, Ning L, Hurst B, Tarbet B, Lin MZ. (2021) Rational design of a new class of protease inhibitors for the potential treatment of coronavirus diseases. bioRxiv, Preprint. DOI: 10.1101/2020.09.15.275891
98. Xia Z, Sacco M, Hu Y, Ma C, Meng X, Zhang F, Szeto T, Xiang Y, Chen Y, Wang J. (2021) Rational Design of Hybrid SARS-CoV-2 Main Protease Inhibitors Guided by the Superimposed Cocrystal Structures with the Peptidomimetic Inhibitors GC-376, Telaprevir, and Boceprevir. ACS Pharmacol Transl Sci, 4 (4): 1408-1421. [PMID:34414360]
99. Yang H, Xie W, Xue X, Yang K, Ma J, Liang W, Zhao Q, Zhou Z, Pei D, Ziebuhr J et al.. (2005) Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol, 3 (10): e324. [PMID:16128623]
100. Yang KS, Ma XR, Ma Y, Alugubelli YR, Scott DA, Vatansever EC, Drelich AK, Sankaran B, Geng ZZ, Blankenship LR et al.. (2020) A Speedy Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors. bioRxiv, Preprint. DOI: 10.1101/2020.07.28.223784 [PMID:32766582]
101. Yang S, Chen SJ, Hsu MF, Wu JD, Tseng CT, Liu YF, Chen HC, Kuo CW, Wu CS, Chang LW et al.. (2006) Synthesis, crystal structure, structure-activity relationships, and antiviral activity of a potent SARS coronavirus 3CL protease inhibitor. J Med Chem, 49 (16): 4971-80. [PMID:16884309]
102. Yin W, Kong WP, Leung SL, Hung CH, Wong KY. (2025) Structure-activity relationship studies of thiazole-based derivatives leading to the identification of novel and potent SARS-CoV-2 main protease inhibitors. Eur J Med Chem, 297: 117952. [PMID:40644922]
103. Zavoronkovs A, Ivanenkov YA, Zagribelnyy B. (2021) Sars-cov-2 inhibitors having covalent modifications for treating coronavirus infections. Patent number: WO2021219089A1. Assignee: Insilico Medicine Ip Limited. Priority date: 30/04/2020. Publication date: 04/11/2021.
104. Zhang C-H, Spasov KA, Reilly RA, Hollander K, Stone EA, Ippolito JA, Liosi M-A, Deshmukh MG, Tirado-Rives J, Zhang S et al.. (2021) Optimization of Triarylpyridinone Inhibitors of the Main Protease of SARS-CoV-2 to Low-Nanomolar Antiviral Potency. ACS Medicinal Chemistry Letters, [Epub ahead of print]. DOI: 10.1021/acsmedchemlett.1c00326
105. Zhang C-H, Stone EA, Deshmukh M, Ippolito JA, Ghahremanpour MM, Tirado-Rives J, Spasov KA, Zhang S, Takeo Y, Kudalkar SN et al.. (2021) Potent Noncovalent Inhibitors of the Main Protease of SARS-CoV-2 from Molecular Sculpting of the Drug Perampanel Guided by Free Energy Perturbation Calculations. ACS Central Science,. DOI: 10.1021/acscentsci.1c00039
106. Zhang CH, Spasov KA, Reilly RA, Hollander K, Stone EA, Ippolito JA, Liosi ME, Deshmukh MG, Tirado-Rives J, Zhang S et al.. (2021) Optimization of Triarylpyridinone Inhibitors of the Main Protease of SARS-CoV-2 to Low-Nanomolar Antiviral Potency. ACS Med Chem Lett, 12 (8): 1325-1332. [PMID:34408808]
107. Zhang CH, Stone EA, Deshmukh M, Ippolito JA, Ghahremanpour MM, Tirado-Rives J, Spasov KA, Zhang S, Takeo Y, Kudalkar SN et al.. (2021) Potent Noncovalent Inhibitors of the Main Protease of SARS-CoV-2 from Molecular Sculpting of the Drug Perampanel Guided by Free Energy Perturbation Calculations. ACS Cent Sci, 7 (3): 467-475. [PMID:33786375]
108. Zhang L, Lin D, Kusov Y, Nian Y, Ma Q, Wang J, von Brunn A, Leyssen P, Lanko K, Neyts J et al.. (2020) α-Ketoamides as Broad-Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, Synthesis, and Activity Assessment. J Med Chem, 63 (9): 4562-4578. [PMID:32045235]
109. Zhang L, Lin D, Sun X, Curth U, Drosten C, Sauerhering L, Backer S, Rox K, Hilgenfeld R. (2020) Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science,. DOI: 10.1126/science.abb3405
110. Zhang L, Lin D, Sun X, Curth U, Drosten C, Sauerhering L, Becker S, Rox K, Hilgenfeld R. (2020) Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science, 368 (6489): 409-412. [PMID:32198291]
111. Zhao Y, Fang C, Zhang Q, Zhang R, Zhao X, Duan Y, Wang H, Zhu Y, Feng L, Zhao J et al.. (2022) Crystal structure of SARS-CoV-2 main protease in complex with protease inhibitor PF-07321332. Protein Cell, 13 (9): 689-693. [PMID:34687004]
112. Zhu W, Xu M, Chen CZ, Guo H, Shen M, Hu X, Shinn P, Klumpp-Thomas C, Michael SG, Zheng W. (2020) Identification of SARS-CoV-2 3CL Protease Inhibitors by a Quantitative High-throughput Screening. ACS Pharmacol Transl Sci, [Epub ahead of print]. DOI: 10.1021/acsptsci.0c00108
113. Zou H, Zhu W, Liu L, Wang W, Xu S, Li Z. (2023) Pyrrolidine antiviral compound. Patent number: WO2023093834A1. Assignee: Xiansheng Zaiming Pharmaceutical Co., Ltd.. Priority date: 25/11/2022. Publication date: 01/06/2023.
114. Identification of inhibitors of SARS-Cov2 M-Pro enzymatic activity using a small molecule repurposing screen. Accessed on 05/11/2024. Modified on 05/11/2024. ChEMBL, https://www.ebi.ac.uk/chembl/web_components/explore/document/CHEMBL4495564. DOI: 10.6019/CHEMBL4495564