Contents:
Previous and Unofficial Names  |
| NendoU | NSP15 |
Database Links  |
|
| ChEMBL Target | CHEMBL4523582 (SARS-CoV-2), CHEMBL5118 (SARS-CoV) |
| UniProtKB | P0DTD1 (SARS-CoV-2), P0C6X7 (SARS-CoV) |
Selected 3D Structures  |
|||||||||||||
|
|
||||||||||||
|
|
||||||||||||
|
|
||||||||||||
Download all structure-activity data for this target as a CSV file 
| Inhibitors | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | View all chemical structures | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| General Comments |
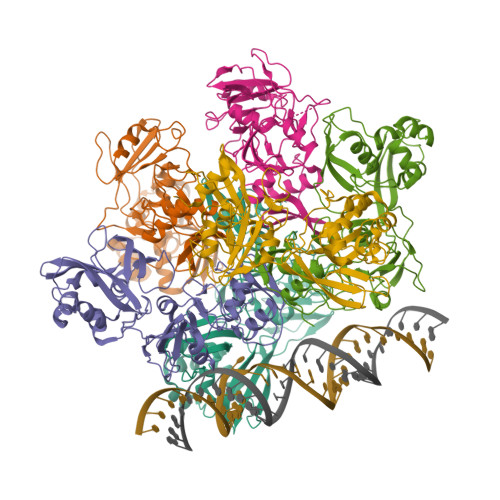
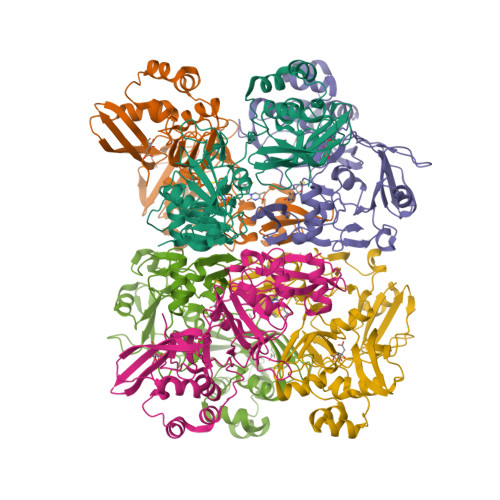
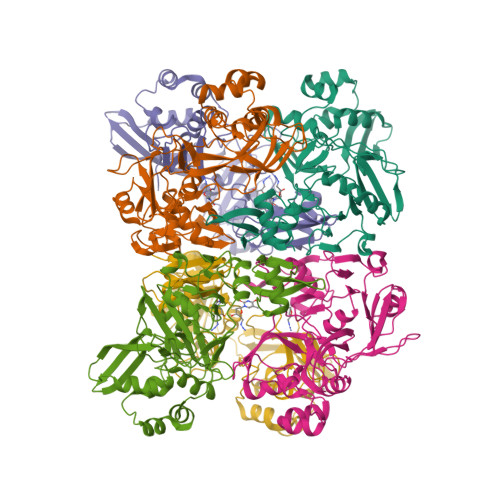
| The coronavirus (CoV) Nsp15 protein is a Mn2+-dependent, uridine specific endoribonuclease that helps the virus avoid triggering a host innate immune response [7,9], primarily by degrading poly(U) RNA (originating from the virus) that would otherwise form double-stranded (ds) RNAs with poly(A) RNA tails and activate host cell dsRNA sensors [1,4,6]. Nsp15 assembles as a homo-hexameric complex [1] that has higher substrate affinity than the enzyme monomer [3]. It is a molecular target for the discovery of new anti-CoV drugs [5,8]. Nsp15 is one of the proteins cleaved from the virus' polyprotein 1ab (pp1ab). We have included it here as a discrete protein to allow curation of interactions with selective chemical modulators. |
References
1. Frazier MN, Dillard LB, Krahn JM, Perera L, Williams JG, Wilson IM, Stewart ZD, Pillon MC, Deterding LJ, Borgnia MJ et al.. (2021) Characterization of SARS2 Nsp15 nuclease activity reveals it's mad about U. Nucleic Acids Res, 49 (17): 10136-10149. [PMID:34403466]
2. Frazier MN, Wilson IM, Krahn JM, Butay KJ, Dillard LB, Borgnia MJ, Stanley RE. (2022) Flipped over U: structural basis for dsRNA cleavage by the SARS-CoV-2 endoribonuclease. Nucleic Acids Res, 50 (14): 8290-8301. [PMID:35801916]
3. Godoy AS, Nakamura AM, Douangamath A, Song Y, Noske GD, Gawriljuk VO, Fernandes RS, Pereira HDM, Oliveira KIZ, Fearon D et al.. (2023) Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease. Nucleic Acids Res, 51 (10): 5255-5270. DOI: 10.1101/2022.09.26.509485 [PMID:37115000]
4. Kim Y, Wower J, Maltseva N, Chang C, Jedrzejczak R, Wilamowski M, Kang S, Nicolaescu V, Randall G, Michalska K et al.. (2021) Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2. Commun Biol, 4 (1): 193. [PMID:33564093]
5. Pakrashy S, Mandal PK, Dey SK, Choudhury SM, Alasmary FA, Almalki AS, Islam MA, Dolai M. (2022) Design of a Structurally Novel Multipotent Drug Candidate by the Scaffold Architecture Technique for ACE-II, NSP15, and Mpro Protein Inhibition: Identification and Isolation of a Natural Product to Prevent the Severity of Future Variants of Covid 19 and a Colorectal Anticancer Drug. ACS Omega, 7 (37): 33408-33422. [PMID:36157758]
6. Pillon MC, Frazier MN, Dillard LB, Williams JG, Kocaman S, Krahn JM, Perera L, Hayne CK, Gordon J, Stewart ZD et al.. (2021) Cryo-EM structures of the SARS-CoV-2 endoribonuclease Nsp15 reveal insight into nuclease specificity and dynamics. Nat Commun, 12 (1): 636. [PMID:33504779]
7. Rashid F, Xie Z, Suleman M, Shah A, Khan S, Luo S. (2022) Roles and functions of SARS-CoV-2 proteins in host immune evasion. Front Immunol, 13: 940756. [PMID:36003396]
8. Sharma A, Kaur M, Yadav P, Singh G, Barnwal RP. (2023) Expediting the drug discovery for ideal leads against SARS-CoV-2 via molecular docking of repurposed drugs. J Biomol Struct Dyn, 41 (16): 7949-7965. [PMID:36165445]
9. Wilson IM, Frazier MN, Li JL, Randall TA, Stanley RE. (2022) Biochemical Characterization of Emerging SARS-CoV-2 Nsp15 Endoribonuclease Variants. J Mol Biol, 434 (20): 167796. [PMID:35995266]









