Contents:
- Gene and Protein Information
- Previous and Unofficial Names
- Database Links
- Selected 3D Structures
- Natural/Endogenous Ligands
- Agonists
- Antagonists
- Transduction Mechanisms
- Tissue Distribution
- Expression Datasets
- Functional Assays
- Physiological Functions
- Physiological Consequences of Altering Gene Expression
- Phenotypes, Alleles and Disease Models
- Gene Expression and Pathophysiology
- Biologically Significant Variants
- References
- Contributors
- How to cite this page
Gene and Protein Information  |
||||||
| class A G protein-coupled receptor | ||||||
| Species | TM | AA | Chromosomal Location | Gene Symbol | Gene Name | Reference |
| Human | 7 | 364 | 9q31.3 | LPAR1 | lysophosphatidic acid receptor 1 | 9 |
| Mouse | 7 | 364 | 4 32.2 cM | Lpar1 | lysophosphatidic acid receptor 1 | 9 |
| Rat | 7 | 364 | 5q24 | Lpar1 | lysophosphatidic acid receptor 1 | 3 |
Database Links  |
|
| Specialist databases | |
| GPCRdb | lpar1_human (Hs), lpar1_mouse (Mm), lpar1_rat (Rn) |
| Other databases | |
| Alphafold | Q92633 (Hs), P61793 (Mm), P61794 (Rn) |
| ChEMBL Target | CHEMBL3819 (Hs), CHEMBL3621025 (Mm), CHEMBL4595 (Rn) |
| Ensembl Gene | ENSG00000198121 (Hs), ENSMUSG00000038668 (Mm), ENSRNOG00000013656 (Rn) |
| Entrez Gene | 1902 (Hs), 14745 (Mm), 116744 (Rn) |
| Human Protein Atlas | ENSG00000198121 (Hs) |
| KEGG Gene | hsa:1902 (Hs), mmu:14745 (Mm), rno:116744 (Rn) |
| OMIM | 602282 (Hs) |
| Pharos | Q92633 (Hs) |
| RefSeq Nucleotide | NM_001401 (Hs), NM_010336 (Mm), NM_053936 (Rn) |
| RefSeq Protein | NP_001392 (Hs), NP_034466 (Mm), NP_446388 (Rn) |
| SynPHARM |
82425 (in complex with ONO-3080573) 82426 (in complex with ONO-9780307) 82427 (in complex with ONO-9910539) |
| UniProtKB | Q92633 (Hs), P61793 (Mm), P61794 (Rn) |
| Wikipedia | LPAR1 (Hs) |
Selected 3D Structures  |
|||||||||||||
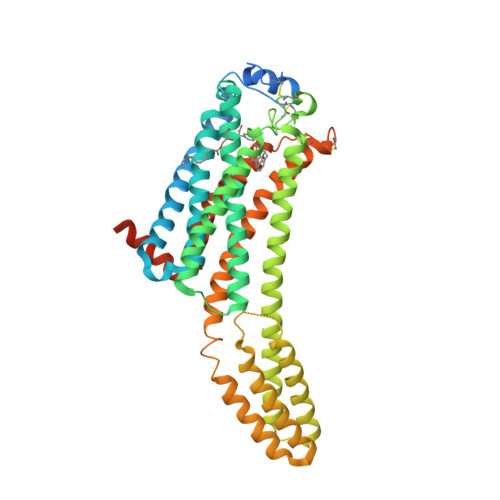
|
|
||||||||||||
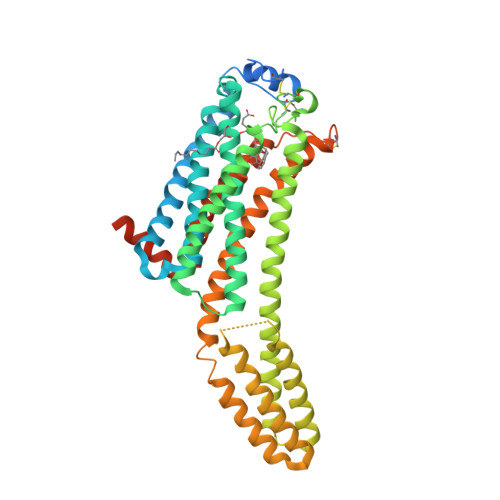
|
|
||||||||||||
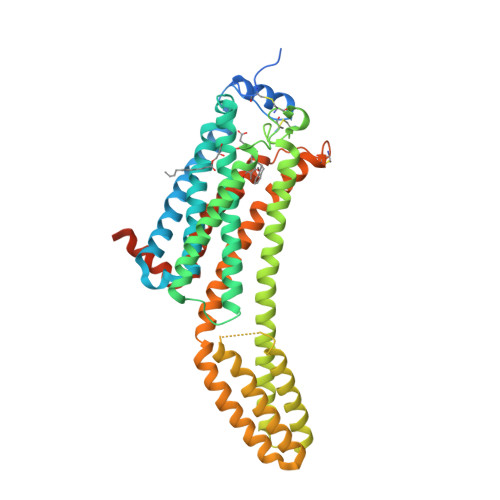
|
|
||||||||||||
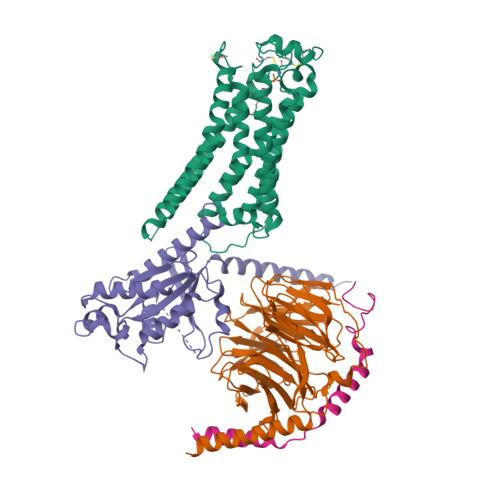
|
|
||||||||||||
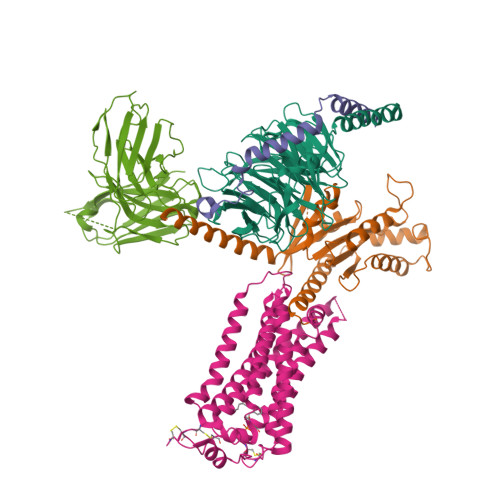
|
|
||||||||||||
Natural/Endogenous Ligands  |
| LPA |
Download all structure-activity data for this target as a CSV file 
| Agonists | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | View all chemical structures | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| View species-specific agonist tables | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Agonist Comments | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Breakdown of affinities for various LPA species [42,44]:
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Antagonists | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | View all chemical structures | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| View species-specific antagonist tables | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Antagonist Comments | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| The IC50 for AM966 [47] and SAR100842 [32] were determined by the LPA-stimulated intracellular calcium release from CHO cells expressing hLPA1 receptors. AM966 inhibited LPA-induced chemotaxis (pIC50 = 6.7) of human IMR-90 lung fibroblasts [47]. SAR100842 inhibited LPA-induced secretion of IL-6, CCL2 and CXCL1 (IC50 ~50 nM) in systemic sclerosis dermal fibroblasts [32]. pKi of AM966 in presence of following LPA species [44]:
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Primary Transduction Mechanisms 
|
|
| Transducer | Effector/Response |
|
Gi/Go family Gq/G11 family G12/G13 family |
Adenylyl cyclase inhibition Phospholipase C stimulation Phospholipase A2 stimulation Other - See Comments |
| Comments: In general LPA1 is known to induce many responses including cell proliferation and survival, cell migration, and cytoskeletal changes; altered cell-cell contact through serum-response element activation, Ca2+ mobilization, and adenylyl cyclase inhibition; and activation of mitogen-activated protein kinase, phospholipase C, Akt, and Rho pathways. For a detailed review please see [7]. | |
| References: 18,29 | |
Tissue Distribution 
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
Expression Datasets  |
|
|
Functional Assays 
|
||||||||||
|
Physiological Functions 
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
Physiological Consequences of Altering Gene Expression 
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
Phenotypes, Alleles and Disease Models 
|
Mouse data from MGI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Expression and Pathophysiology Comments | |
| Mouse models indicate that overactivation of LPA1 during fetal development initiates hydrocephalus in vivo [53], and can cause prenatal intracerebral hemorrhage leading to schizophrenia-like brain and behavioral changes [36]. Also in mice, fetal hypoxia activates LPA1 signaling, inhibition of which reduces hypoxia-induced brain injury. These findings may point to potential treatments for fetal hypoxia-induced CNS disorders or for other forms of hypoxic brain injury [27]. |
| Biologically Significant Variant Comments |
| There is a splice variant (mrec 1.3) that results in an 18-amino acid deletion of the N terminus, but its biological significance is unknown [9]. |
References
1. Akasaka H, Tanaka T, Sano FK, Matsuzaki Y, Shihoya W, Nureki O. (2022) Structure of the active Gi-coupled human lysophosphatidic acid receptor 1 complexed with a potent agonist. Nat Commun, 13 (1): 5417. [PMID:36109516]
2. Alioli CA, Demesmay L, Laurencin-Dalacieux S, Beton N, Farlay D, Follet H, Saber A, Duboeuf F, Chun J, Rivera R et al.. (2020) Expression of the type 1 lysophosphatidic acid receptor in osteoblastic cell lineage controls both bone mineralization and osteocyte specification. Biochim Biophys Acta Mol Cell Biol Lipids, 1865 (8): 158715. [PMID:32330664]
3. Allard J, Barrón S, Diaz J, Lubetzki C, Zalc B, Schwartz JC, Sokoloff P. (1998) A rat G protein-coupled receptor selectively expressed in myelin-forming cells. Eur J Neurosci, 10 (3): 1045-53. [PMID:9753172]
4. An S, Bleu T, Hallmark OG, Goetzl EJ. (1998) Characterization of a novel subtype of human G protein-coupled receptor for lysophosphatidic acid. J Biol Chem, 273 (14): 7906-10. [PMID:9525886]
5. Bandoh K, Aoki J, Taira A, Tsujimoto M, Arai H, Inoue K. (2000) Lysophosphatidic acid (LPA) receptors of the EDG family are differentially activated by LPA species. Structure-activity relationship of cloned LPA receptors. FEBS Lett, 478 (1-2): 159-65. [PMID:10922489]
6. Cheng PTW, Kaltenbach 3rd RF, Zhang H, Shi J, Tao S, Li J, Kennedy LJ, Walker SJ, Shi Y, Wang Y et al.. (2021) Discovery of an Oxycyclohexyl Acid Lysophosphatidic Acid Receptor 1 (LPA1) Antagonist BMS-986278 for the Treatment of Pulmonary Fibrotic Diseases. J Med Chem, 64 (21): 15549-15581. [PMID:34709814]
7. Choi JW, Herr DR, Noguchi K, Yung YC, Lee CW, Mutoh T, Lin ME, Teo ST, Park KE, Mosley AN, Chun J. (2010) LPA receptors: subtypes and biological actions. Annu Rev Pharmacol Toxicol, 50: 157-86. [PMID:20055701]
8. Chrencik JE, Roth CB, Terakado M, Kurata H, Omi R, Kihara Y, Warshaviak D, Nakade S, Asmar-Rovira G, Mileni M et al.. (2015) Crystal Structure of Antagonist Bound Human Lysophosphatidic Acid Receptor 1. Cell, 161 (7): 1633-43. [PMID:26091040]
9. Contos JJ, Chun J. (1998) Complete cDNA sequence, genomic structure, and chromosomal localization of the LPA receptor gene, lpA1/vzg-1/Gpcr26. Genomics, 51 (3): 364-78. [PMID:9721207]
10. Contos JJ, Fukushima N, Weiner JA, Kaushal D, Chun J. (2000) Requirement for the lpA1 lysophosphatidic acid receptor gene in normal suckling behavior. Proc Natl Acad Sci USA, 97 (24): 13384-9. [PMID:11087877]
11. Dancs PT, Ruisanchez É, Balogh A, Panta CR, Miklós Z, Nüsing RM, Aoki J, Chun J, Offermanns S, Tigyi G et al.. (2017) LPA1 receptor-mediated thromboxane A2 release is responsible for lysophosphatidic acid-induced vascular smooth muscle contraction. FASEB J, 31 (4): 1547-1555. [PMID:28069828]
12. Dubin AE, Bahnson T, Weiner JA, Fukushima N, Chun J. (1999) Lysophosphatidic acid stimulates neurotransmitter-like conductance changes that precede GABA and L-glutamate in early, presumptive cortical neuroblasts. J Neurosci, 19 (4): 1371-81. [PMID:9952414]
13. Dubin AE, Herr DR, Chun J. (2010) Diversity of lysophosphatidic acid receptor-mediated intracellular calcium signaling in early cortical neurogenesis. J Neurosci, 30 (21): 7300-9. [PMID:20505096]
14. Durgam GG, Virag T, Walker MD, Tsukahara R, Yasuda S, Liliom K, van Meeteren LA, Moolenaar WH, Wilke N, Siess W et al.. (2005) Synthesis, structure-activity relationships, and biological evaluation of fatty alcohol phosphates as lysophosphatidic acid receptor ligands, activators of PPARgamma, and inhibitors of autotaxin. J Med Chem, 48 (15): 4919-30. [PMID:16033271]
15. Estivill-Torrús G, Llebrez-Zayas P, Matas-Rico E, Santín L, Pedraza C, De Diego I, Del Arco I, Fernández-Llebrez P, Chun J, De Fonseca FR. (2008) Absence of LPA1 signaling results in defective cortical development. Cereb Cortex, 18 (4): 938-50. [PMID:17656621]
16. Fischer DJ, Nusser N, Virag T, Yokoyama K, Wang Da, Baker DL, Bautista D, Parrill AL, Tigyi G. (2001) Short-chain phosphatidates are subtype-selective antagonists of lysophosphatidic acid receptors. Mol Pharmacol, 60 (4): 776-84. [PMID:11562440]
17. Fukushima N, Ishii I, Habara Y, Allen CB, Chun J. (2002) Dual regulation of actin rearrangement through lysophosphatidic acid receptor in neuroblast cell lines: actin depolymerization by Ca(2+)-alpha-actinin and polymerization by rho. Mol Biol Cell, 13 (8): 2692-705. [PMID:12181339]
18. Fukushima N, Kimura Y, Chun J. (1998) A single receptor encoded by vzg-1/lpA1/edg-2 couples to G proteins and mediates multiple cellular responses to lysophosphatidic acid. Proc Natl Acad Sci USA, 95 (11): 6151-6. [PMID:9600933]
19. Fukushima N, Shano S, Moriyama R, Chun J. (2007) Lysophosphatidic acid stimulates neuronal differentiation of cortical neuroblasts through the LPA1-G(i/o) pathway. Neurochem Int, 50 (2): 302-7. [PMID:17056154]
20. Fukushima N, Weiner JA, Chun J. (2000) Lysophosphatidic acid (LPA) is a novel extracellular regulator of cortical neuroblast morphology. Dev Biol, 228 (1): 6-18. [PMID:11087622]
21. Fukushima N, Weiner JA, Kaushal D, Contos JJ, Rehen SK, Kingsbury MA, Kim KY, Chun J. (2002) Lysophosphatidic acid influences the morphology and motility of young, postmitotic cortical neurons. Mol Cell Neurosci, 20 (2): 271-82. [PMID:12093159]
22. González-Gil I, Zian D, Vázquez-Villa H, Hernández-Torres G, Martínez RF, Khiar-Fernández N, Rivera R, Kihara Y, Devesa I, Mathivanan S et al.. (2020) A Novel Agonist of the Type 1 Lysophosphatidic Acid Receptor (LPA1), UCM-05194, Shows Efficacy in Neuropathic Pain Amelioration. J Med Chem, 63 (5): 2372-2390. [PMID:31790581]
23. Guillot E, Le Bail JC, Paul P, Fourgous V, Briand P, Partiseti M, Cornet B, Janiak P, Philippo C. (2020) Lysophosphatidic Acid Receptor Agonism: Discovery of Potent Nonlipid Benzofuran Ethanolamine Structures. J Pharmacol Exp Ther, 374 (2): 283-294. [PMID:32409422]
24. Heasley BH, Jarosz R, Lynch KR, Macdonald TL. (2004) Initial structure-activity relationships of lysophosphatidic acid receptor antagonists: discovery of a high-affinity LPA1/LPA3 receptor antagonist. Bioorg Med Chem Lett, 14 (11): 2735-40. [PMID:15125924]
25. Hecht JH, Weiner JA, Post SR, Chun J. (1996) Ventricular zone gene-1 (vzg-1) encodes a lysophosphatidic acid receptor expressed in neurogenic regions of the developing cerebral cortex. J Cell Biol, 135 (4): 1071-83. [PMID:8922387]
26. Heise CE, Santos WL, Schreihofer AM, Heasley BH, Mukhin YV, Macdonald TL, Lynch KR. (2001) Activity of 2-substituted lysophosphatidic acid (LPA) analogs at LPA receptors: discovery of a LPA1/LPA3 receptor antagonist. Mol Pharmacol, 60 (6): 1173-80. [PMID:11723223]
27. Herr KJ, Herr DR, Lee CW, Noguchi K, Chun J. (2011) Stereotyped fetal brain disorganization is induced by hypoxia and requires lysophosphatidic acid receptor 1 (LPA1) signaling. Proc Natl Acad Sci USA, 108 (37): 15444-9. [PMID:21878565]
28. Im DS. (2010) Pharmacological tools for lysophospholipid GPCRs: development of agonists and antagonists for LPA and S1P receptors. Acta Pharmacol Sin, 31 (9): 1213-22. [PMID:20729877]
29. Ishii I, Contos JJ, Fukushima N, Chun J. (2000) Functional comparisons of the lysophosphatidic acid receptors, LP(A1)/VZG-1/EDG-2, LP(A2)/EDG-4, and LP(A3)/EDG-7 in neuronal cell lines using a retrovirus expression system. Mol Pharmacol, 58 (5): 895-902. [PMID:11040035]
30. Kano K, Arima N, Ohgami M, Aoki J. (2008) LPA and its analogs-attractive tools for elucidation of LPA biology and drug development. Curr Med Chem, 15 (21): 2122-31. [PMID:18781939]
31. Kingsbury MA, Rehen SK, Contos JJ, Higgins CM, Chun J. (2003) Non-proliferative effects of lysophosphatidic acid enhance cortical growth and folding. Nat Neurosci, 6 (12): 1292-9. [PMID:14625558]
32. Ledein L, Léger B, Dees C, Beyer C, Distler A, Vettori S, Boukaiba R, Bidouard JP, Schaefer M, Pernerstorfer J et al.. (2020) Translational engagement of lysophosphatidic acid receptor 1 in skin fibrosis: from dermal fibroblasts of patients with scleroderma to tight skin 1 mouse. Br J Pharmacol, 177 (18): 4296-4309. [PMID:32627178]
33. Liu S, Paknejad N, Zhu L, Kihara Y, Ray M, Chun J, Liu W, Hite RK, Huang XY. (2022) Differential activation mechanisms of lipid GPCRs by lysophosphatidic acid and sphingosine 1-phosphate. Nat Commun, 13 (1): 731. [PMID:35136060]
34. Lummis NC, Sánchez-Pavón P, Kennedy G, Frantz AJ, Kihara Y, Blaho VA, Chun J. (2019) LPA1/3 overactivation induces neonatal posthemorrhagic hydrocephalus through ependymal loss and ciliary dysfunction. Sci Adv, 5 (10): eaax2011. [PMID:31633020]
35. Matas-Rico E, García-Diaz B, Llebrez-Zayas P, López-Barroso D, Santín L, Pedraza C, Smith-Fernández A, Fernández-Llebrez P, Tellez T, Redondo M et al.. (2008) Deletion of lysophosphatidic acid receptor LPA1 reduces neurogenesis in the mouse dentate gyrus. Mol Cell Neurosci, 39 (3): 342-55. [PMID:18708146]
36. Mirendil H, Thomas EA, De Loera C, Okada K, Inomata Y, Chun J. (2015) LPA signaling initiates schizophrenia-like brain and behavioral changes in a mouse model of prenatal brain hemorrhage. Transl Psychiatry, 5: e541. [PMID:25849980]
37. Ohta H, Sato K, Murata N, Damirin A, Malchinkhuu E, Kon J, Kimura T, Tobo M, Yamazaki Y, Watanabe T, Yagi M, Sato M, Suzuki R, Murooka H, Sakai T, Nishitoba T, Im DS, Nochi H, Tamoto K, Tomura H, Okajima F. (2003) Ki16425, a subtype-selective antagonist for EDG-family lysophosphatidic acid receptors. Mol Pharmacol, 64 (4): 994-1005. [PMID:14500756]
38. Ohuchi H, Hamada A, Matsuda H, Takagi A, Tanaka M, Aoki J, Arai H, Noji S. (2008) Expression patterns of the lysophospholipid receptor genes during mouse early development. Dev Dyn, 237 (11): 3280-94. [PMID:18924241]
39. Olianas MC, Dedoni S, Onali P. (2020) Antidepressants induce profibrotic responses via the lysophosphatidic acid receptor LPA1. Eur J Pharmacol, 873: 172963. [PMID:32007501]
40. Pedraza C, Sánchez-López J, Castilla-Ortega E, Rosell-Valle C, Zambrana-Infantes E, García-Fernández M, Rodriguez de Fonseca F, Chun J, Santín LJ, Estivill-Torrús G. (2014) Fear extinction and acute stress reactivity reveal a role of LPA(1) receptor in regulating emotional-like behaviors. Brain Struct Funct, 219 (5): 1659-72. [PMID:23775489]
41. Qian L, Xu Y, Simper T, Jiang G, Aoki J, Umezu-Goto M, Arai H, Yu S, Mills GB, Tsukahara R et al.. (2006) Phosphorothioate analogues of alkyl lysophosphatidic acid as LPA3 receptor-selective agonists. ChemMedChem, 1 (3): 376-83. [PMID:16892372]
42. Ray M, Nagai K, Kihara Y, Kussrow A, Kammer MN, Frantz A, Bornhop DJ, Chun J. (2020) Unlabeled lysophosphatidic acid receptor binding in free solution as determined by a compensated interferometric reader. J Lipid Res, 61 (8): 1244-1251. [PMID:32513900]
43. Rivera RR, Lin ME, Bornhop EC, Chun J. (2020) Conditional Lpar1 gene targeting identifies cell types mediating neuropathic pain. FASEB J, 34 (7): 8833-8842. [PMID:32929779]
44. Sattikar A, Dowling MR, Rosethorne EM. (2017) Endogenous lysophosphatidic acid (LPA1 ) receptor agonists demonstrate ligand bias between calcium and ERK signalling pathways in human lung fibroblasts. Br J Pharmacol, 174 (3): 227-237. [PMID:27864940]
45. Simon MF, Daviaud D, Pradère JP, Grès S, Guigné C, Wabitsch M, Chun J, Valet P, Saulnier-Blache JS. (2005) Lysophosphatidic acid inhibits adipocyte differentiation via lysophosphatidic acid 1 receptor-dependent down-regulation of peroxisome proliferator-activated receptor gamma2. J Biol Chem, 280 (15): 14656-62. [PMID:15710620]
46. Swaney JS, Chapman C, Correa LD, Stebbins KJ, Broadhead AR, Bain G, Santini AM, Darlington J, King CD, Baccei CS et al.. (2011) Pharmacokinetic and pharmacodynamic characterization of an oral lysophosphatidic acid type 1 receptor-selective antagonist. J Pharmacol Exp Ther, 336 (3): 693-700. [PMID:21159750]
47. Swaney JS, Chapman C, Correa LD, Stebbins KJ, Bundey RA, Prodanovich PC, Fagan P, Baccei CS, Santini AM, Hutchinson JH et al.. (2010) A novel, orally active LPA(1) receptor antagonist inhibits lung fibrosis in the mouse bleomycin model. Br J Pharmacol, 160 (7): 1699-713. [PMID:20649573]
48. Terakado M, Suzuki H, Hashimura K, Tanaka M, Ueda H, Kohno H, Fujimoto T, Saga H, Nakade S, Habashita H et al.. (2016) Discovery of ONO-7300243 from a Novel Class of Lysophosphatidic Acid Receptor 1 Antagonists: From Hit to Lead. ACS Med Chem Lett, 7 (10): 913-918. [PMID:27774128]
49. Ueda H, Neyama H, Matsushita Y. (2020) Lysophosphatidic Acid Receptor 1- and 3-Mediated Hyperalgesia and Hypoalgesia in Diabetic Neuropathic Pain Models in Mice. Cells, 9 (8). [PMID:32824296]
50. Weiner JA, Chun J. (1999) Schwann cell survival mediated by the signaling phospholipid lysophosphatidic acid. Proc Natl Acad Sci USA, 96 (9): 5233-8. [PMID:10220449]
51. Weiner JA, Fukushima N, Contos JJ, Scherer SS, Chun J. (2001) Regulation of Schwann cell morphology and adhesion by receptor-mediated lysophosphatidic acid signaling. J Neurosci, 21 (18): 7069-78. [PMID:11549717]
52. Yang AH, Ishii I, Chun J. (2002) In vivo roles of lysophospholipid receptors revealed by gene targeting studies in mice. Biochim Biophys Acta, 1582 (1-3): 197-203. [PMID:12069829]
53. Yung YC, Mutoh T, Lin ME, Noguchi K, Rivera RR, Choi JW, Kingsbury MA, Chun J. (2011) Lysophosphatidic acid signaling may initiate fetal hydrocephalus. Sci Transl Med, 3 (99): 99ra87. [PMID:21900594]
54. Zhang H, Xu X, Gajewiak J, Tsukahara R, Fujiwara Y, Liu J, Fells JI, Perygin D, Parrill AL, Tigyi G et al.. (2009) Dual activity lysophosphatidic acid receptor pan-antagonist/autotaxin inhibitor reduces breast cancer cell migration in vitro and causes tumor regression in vivo. Cancer Res, 69 (13): 5441-9. [PMID:19509223]












